Abstract
Reprogramming somatic cells to pluripotent stem cells has revolutionized the biomedical field by providing enormous hopes and opportunities for the regeneration of tissues and organs for transplantation. Using a small molecule cocktail of epigenetic modifiers and cell signalling inhibitors, a chemical-based easy and controllable technique for converting human somatic cells into chemically induced pluripotent stem cells was recently reported (Guan, Nature 605:325–31, 2022). This novel approach offers well-defined, safe, simple, easy, and clinical-grade manufacturing strategies for modifying the fate of human cells required for regenerative therapeutics.
Similar content being viewed by others
Main text
Since Yamanaka’s team identified four key factors in 2006–2007 (Takahashi and Yamanaka 2006), researchers have been looking for a chemical cocktail to reduce the viral load involved in somatic cell reprogramming ever since. Episomal, RNP, protein-based, and miRNA techniques are among the others that have been documented, but each has its drawbacks and can only reprogram cells with an efficiency of 0.01 to 1% (al Abbar et al. 2020). Chemical inducers that target cell signalling pathways and epigenetic modifiers have been shown to reprogram mouse somatic cells and transdifferentiate (also known as lineage reprogram) human and mouse cells. Despite this, previous attempts to reprogram human somatic cells have failed, because these cells maintain a stable epigenome (Takeda et al. 2018; Yang et al. 2022; Xu, Du, and Deng 2015).
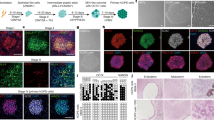
Following leads from mouse somatic cell chemical reprogramming and chemical library screens, (Guan et al., 2022) the authors reported a cocktail of 16 small molecules consisting of CHIR99021, 616,452, TTNPB, Y27632, ABT869, SAG, JNKIN8, 5-azacytidine, tranylcypromine, valproic acid, DZNep, EPZ004777, UNC0379, and PD0325901 required in stage-specific manner for conversion of fibroblasts to epithelial-like cells and later for activating pluripotency master regulator, Oct 4. The colonies obtained on chemical reprogramming (designated as human chemically iPS cells (hCiPS)) displayed typical hES cell morphology, doubling time, transcriptomic and epigenomic profiles.
The authors used a similar chemical reprogramming approach on adult somatic cells such as adipose-derived mesenchymal stromal cells and adult skin dermal fibroblasts to generate hCiPS with efficiencies ranging from 0.21 ± 0.07% to 2.56 ± 0.63%. Adult somatic-cell-derived hCiPS cells were able to produce cell types from three germ layers in vivo and in vitro, and their transcriptome and epigenetic profiles were similar to those of hES cells.
During the reprogramming trajectory in the current study using human cells, various cell states known as an intermediate plastic state, an extraembryonic endoderm-like (XEN-like) state, and naive pluripotency state were identified, as previously reported from the same group in mouse cells (Zhao et al. 2015; Li et al. 2017). In contrast, primitive streak-like intermediates are observed in OSKM-induced reprogramming, whereas the XEN-like state is exclusive to chemical reprogramming and is rarely documented (Parenti et al. 2016). Interestingly, authors reported that during the intermediate plastic state, cells were reprogrammed to acquire characteristics of developing human limb bud cells that are similar to amphibian (axolotl) limb regeneration, where regeneration-like gene regulatory programs governing embryonic limb development are reactivated which is indispensable for acquiring cell pluripotency.
In contrast to studies of direct chemical transdifferentiation, in which cells do not transition to any transitory pluripotent state, the current investigation highlighted the establishment of an intermediate plastic population that resembles genes relevant to limb development across species. This plastic state is reported to have open chromatin and reduced DNA methylation at a global level.
One of the limitations of CiPS cells is the longer duration of reprogramming of 5–6 weeks vs 2–4 weeks for viral and episomal approaches. However, more comprehensive analysis, and additional optimizations are required to test the proposed cocktail with other somatic cells from humans and different species for reprogramming efficiency. Along similar lines, a recent report describes the use of three small molecules alone to generate ‘totipotent’ mouse cells from pluripotent stem cells (Hu et al. 2022). Furthermore, more insight is required to determine whether the small molecules used have potential off target effects prior to evaluating their clinical implications. Future research on retention and the role of epigenetic memory in chemical reprogramming would open further avenues for the regenerative field by providing an opportunity to select appropriate starting cell material for therapies.
Conclusions
It is intriguing to note that the authors of the current study employed a chemical cocktail that inhibits global DNMT methylation, histone acetylation, histone methylation, and other major cell signalling pathways, which in turn provides an open (environment) chromatin accessible to a variety of transcription factors and erases the cell’s history in order to write a new paradigm. The proposed approach has significant promise for stimulating in vivo repair and regeneration of patients’ endogenous cells. Furthermore, the method minimizes the risk of tumorigenesis associated with viral methods (Fig. 1).
Availability of data and materials
Not applicable.
References
Al Abbar A, Ngai SC, Nograles N, Alhaji SY, Abdullah S. Induced pluripotent stem cells: reprogramming platforms and applications in cell replacement therapy. BioResearch Open Access. 2020;9(1):121–36. https://doi.org/10.1089/biores.2019.0046.
Guan J, Wang G, Wang J, Zhang Z, Yao F, Cheng L, et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature. 2022;605(7909):325–31. https://doi.org/10.1038/s41586-022-04593-5.
Hu Y, Yang Y, Tan P, Zhang Y, Han M, Jiawei Y, et al. Induction of mouse totipotent stem cells by a defined chemical cocktail. Nature. 2022. https://doi.org/10.1038/s41586-022-04967-9.
Li X, Liu D, Ma Y, Xiaomin D, Jing J, Wang L, et al. Direct reprogramming of fibroblasts via a chemically induced XEN-like state. Cell Stem Cell. 2017;21(2):264–273.e7. https://doi.org/10.1016/j.stem.2017.05.019.
Parenti A, Halbisen MA, Wang K, Latham K, Ralston A. OSKM induce extraembryonic endoderm stem cells in parallel to induced pluripotent stem cells. Stem Cell Reports. 2016;6(4):447–55. https://doi.org/10.1016/j.stemcr.2016.02.003.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. https://doi.org/10.1016/j.cell.2006.07.024.
Takeda Y, Harada Y, Yoshikawa T, Dai P. Chemical compound-based direct reprogramming for future clinical applications. Biosci Rep. 2018;38(3):BSR20171650. https://doi.org/10.1042/BSR20171650.
Xu J, Yuanyuan D, Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16(2):119–34. https://doi.org/10.1016/j.stem.2015.01.013.
Yang Z, Xiaochan X, Chan G, Nielsen AV, Chen G, Guo F, et al., editors. Chemical pretreatment activated a plastic state amenable to direct lineage reprogramming. Front Cell Dev Biol. 2022;10:865038. https://doi.org/10.3389/fcell.2022.865038.
Zhao Y, Zhao T, Jingyang Guan X, Zhang YF, Ye J, Zhu J, et al. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell. 2015;163(7):1678–91. https://doi.org/10.1016/j.cell.2015.11.017.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
D.A. conceptualized the idea, wrote and finalized the manuscript. The author(s) read and approved the final manuscript.
Authors’ information
Deepri Abbey, PhD is currently working as a Head-Stem cell Core at inStem Bangalore (India). The Core aims at providing training, services, and resources to investigators to facilitate stem cell research.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Abbey, D. Chemical journey of somatic cells to pluripotency. Cell Regen 11, 27 (2022). https://doi.org/10.1186/s13619-022-00126-7
Published:
DOI: https://doi.org/10.1186/s13619-022-00126-7