Abstract
Endophytic bacteria are one of the symbiotic microbial groups closely related to host algae. However, less research on the endophytic bacteria of marine algae. In this study, the endophytic bacterial community of Sargassum thunbergii was investigated using the culture method and high-throughput sequencing. Thirty-nine endophytic bacterial strains, belonging to two phyla, five genera and sixteen species, were isolated, and Firmicutes, Bacillus and Metabacillus indicus were the dominant taxa at the phylum, genus and species level, respectively. High-throughput sequencing revealed 39 phyla and 574 genera of endophytic bacteria, and the dominant phylum was Proteobacteria, while the dominant genus was Ralstonia. The results also indicated that the endophytic bacteria of S. thunbergii included various groups with nitrogen fixation, salt tolerance, pollutant degradation, and antibacterial properties but also contained some pathogenic bacteria. Additionally, the endophytic bacterial community shared a large number of groups with the epiphytic bacteria and bacteria in the surrounding seawater, but the three groups of samples could be clustered separately. In conclusion, there are a variety of functional endophytic bacteria living in S. thunbergii, and the internal condition of algae is a selective factor for the formation of endophytic bacterial communities. This study enriched the database of endophytic bacteria in marine macroalgae, paving the way for further understanding of the interrelationships between endophytic bacteria, macroalgae, and the environment.
Similar content being viewed by others
Introduction
Endophytic bacteria are an important component of plant endophytic microorganisms and play an important role in the growth, development and disease resistance of host plants (Wang et al. 2015; Lu et al. 2022). The study of endophytic bacterial diversity not only provides valuable microbial resources, but also has important value in improving plant yield and elucidating the mechanisms of plant resistance and adaptability. Current research on plant endophytic bacteria is focused on higher terrestrial plants, especially agricultural crops, medicinal plants and environmental remediation plants. Numerous studies on the isolation, identification and determination of the physiological activity of endophytic bacteria from a wide range of terrestrial plants (Zhang et al. 2023; Purushotham et al. 2020) along with aquatic plant (Lan et al. 2008) and marine plants (Li et al. 2017; Zhang et al. 2021a; Ran et al. 2016) have been conducted since the middle of the 20th century.
Marine macroalgae, the main primary producers in coastal ecosystems, play an important ecological role by providing food, shelter, and habitat for many marine organisms, and having high industrial, agricultural, edible and medicinal value (Melo et al. 2021; Subramanian and Marudhamuthu 2020). However, only a few studies have also been carried out on the endophytic bacteria of marine macroalgae, mainly on some physiological activities of the isolated endophytic strains, such as the phosphorus-dissolving ability of the endophytic bacteria isolated from Myriophyllum spicatum (Guo et al. 2016), the resistance and tolerance of the endophytic bacteria of Bacillus and Acinetobacter to manganese stress, the growth-promoting ability of host plants isolated from wild and cultured Myriophyllum verticillatum (Zhang 2017), and the taxol-producing endophytic bacteria from Sargassum polycystum and Acanthaphora specifera (Subramanian and Marudhamuthu 2020), as well as the antibacterial activity of the endophytic bacteria isolated from Acanthophora dendroides, Sargassum sabrepandum (Ahmed et al. 2016) and Eucheuma spinosum (Sugrani et al. 2019), and the antioxidant activity of the endophytic bacteria isolated from Ulva lactuca L. and Undaria pinnatifida Suringar (Feng et al. 2020), etc.
Most of the above studies are based on the traditional cultivation method. High-throughput sequencing has been widely used to determine the endophytic bacterial communities of various higher plants, which can comprehensively clarify the structure and diversity of microbial communities. It is found that some dominant phyla are the same in the endophytic bacterial communities of different plants, but there are great differences at the genus level. For example, the three main phyla of the endophytic bacteria in sweet potato Ipomoea batatas (Puri et al. 2019) were found to be Proteobacteria, Bacteroidetes and Actinomycetes, similar to the dominant phyla of faba bean (Vicia faba L) seeds (Bacteroidetes, Proteobacteria, Firmicutes and Actinomycetes) (Liu et al. 2021b) and Polygonatum cyrtonema (Proteobacteria, Actinomycetes, Gemmatimonadetes, Firmicutes and Acidobacteria) (Cai et al. 2020). This is also very similar to the dominant endophytic bacteria in the aquatic plant Spirodela polyrhiza (Proteobacteria, Bacteroides, Actinomycetes and Acidobacteria) (Li et al. 2021). However, according to the results of high-throughput sequencing, the endophytic bacteria at the genus level vary greatly among species. For example, the core bacteria at the genus level in Polygonatum are composed of Sphingomonas, Gemmatimonas and Streptomyces (Cai et al. 2020). However, the endophytic bacteria at the genus level in S. polyrhiza are mainly some genera that are closely related to the biogeochemical cycles of nitrogen, carbon and phosphorus in water bodies, including Pelomonas, Flavobacterium, Rubrivivax, Hydrogenophaga and Sphingomonas (Li et al. 2021). Moreover, for the first time, many taxa have been reported as endophytic bacteria, such as the endophytic bacterial community found in rice growing in saline soil in West Bengal along with some previously unreported endophytic bacteria, such as Aerinimonas, Arcobacter, Chitinophaga, Hydroispora and Sulfospirillum (Kunda et al. 2018).
However, little is known about the structure of the endophytic bacterial communities in marine algae. only Hollants et al. (Hollants et al. 2011) have conducted a study about the endophytic bacterial communities in green seaweed Bryopsis (Bryopsidales, Chlorophyta) using the DGGE (denaturing gradient electrophoresis) method, and Mei analyzed the composition of endophytic bacterial community in Sargassum horneri and Ulva prolifera (Mei 2019) based the high-throughput sequencing. Additionally, Mangun et al. compared the isolated endophytic bacteria from healthy and diseased Kappahycus alvarezii (Mangun et al. 2023) Determining the composition and diversity of the endophytic bacterial communities in marine macroalgae using high-throughput sequencing technology is the first issue we want to investigate.
Additionally, endophytic bacteria in plants are closely related to their hosts and environment. Only a small proportion of endophytic bacteria in plants are unique to the plant and undergo vertical migration from parent to offspring through reproductive organs such as seeds or pollen. However, most of the bacteria in the whole plant are introduced from outside during plant growth (Wang et al. 2015). Many studies have shown that the composition and structure of epiphytic bacteria in marine macroalgae were different from those in surrounding waters (Mancuso et al. 2016; Pei et al. 2021), and it has also been found that the epiphytic and endophytic bacterial communities of Gracilaria dura were included in a sub-population of bacteria (Singh et al. 2015), However, few studies have simultaneously studied both endophytic and epiphytic bacterial communities and bacterial communities in the seawater surrounding macroalgae. The difference between the endophytic bacterial community in macroalgae, the epiphytic bacteria on the surface of macroalgae and the bacterial community in the seawater around macroalgae is another issue of interest to us.
Sargassum thunbergii is a common intertidal macroalgae along the northern coast of China, which has great value in medicine, the chemical industry, aquaculture and marine ecosystem restoration (Wu et al. 2010; Wang et al. 2022), and large-scale artificial cultures of this alga have been carried out. In this study, the community structure and diversity of the endophytic bacteria in S. thunbergii from the intertidal zone of Qingdao, Shandong Peninsula (China), were investigated using the culture method and high-throughput sequencing technology. At the same time, we compared their similarities and differences with the algal epiphytic bacteria and the bacterial community in the surrounding seawater. The purpose of our study was to broaden the basic information of marine algal endophytic bacterial communities and obtain culturable endophytic bacterial for further research. Our research helps to understand the relationship between algal endophytic bacteria, epiphytic bacteria and bacteria in the surrounding seawater and provides an experimental and theoretical basis for the development and utilization of S. thunbergii.
Materials and methods
Sample collection and processing
Well-grown S. thunbergii samples were collected from the rocky intertidal zone of Taipingjiao, Qingdao, Shandong Province (36°14’58.3"N, 120°21’34.2"E), in November 2020 and placed in sterile sample bags. The surrounding seawater was also collected in pre-sterilized sample bottles. Both the algal and seawater samples were stored in a portable ice box and transferred to the laboratory within 30 min for further processing.
Sample surface disinfection
The algal samples were rinsed with sterile seawater to remove loosely adhering epiphytes and sand, and then the surface water of S. thunbergii was blotted out with sterile filter paper. In the pre-experiment, the samples were submerged in 75% ethanol for 5 min and in 2.5% sodium hypochlorite for 10 min successively, and then washed seven times with sterile double-distilled water (Liu et al. 2022). The final rinse was collected and incubated on Zobell 2216E agar medium plates at 25 °C in DNP-9082 Constant temperature incubator (Jinghong Experimental instrument Co., Ltd, Shanghai) for 72 h; when no bacterial growth was observed on the plate, we deemed the disinfection to be successful (Benjelloun et al. 2019; de Jong et al. 2019).
Culture method
(1) Cultivation, isolation and preservation of endophytic bacteria.
An amount of 2.0 g of disinfected S. thunbergii was placed in a sterile mortar, adding 8 mL sterile physiological saline, which was then grinded to obtain a homogenous suspension. The initial suspension and decimal dilutions were used to isolate bacteria on Zobell 2216E agar medium plates. Three samples were treated, and the plates were incubated at 28 °C in DNP-9082 Constant temperature incubator (Jinghong Experimental instrument Co., Ltd, Shanghai) for 48 h. All colonies on a plate spread with one of the appropriate dilutions were isolated and purified several times, and then the obtained bacterial strains were preserved with cryoprotectant (0.9% physiological saline, 15% glycerol) (Peng et al. 2020) at -80 °C for further research.
(2) DNA extraction of endophytic bacteria.
Bacterial DNA was extracted using a bacterial genomic DNA extraction kit (TIANamp Bacteria DNA Kit), operated according to the kit instructions; only the second step was changed by adding 180 µL of 50 mg/mL lysozyme buffer with treatment at 37 °C for more than 30 min, while the other steps remained unchanged.
(3) Identification of endophytic bacteria via 16S rDNA sequencing.
After DNA extraction, sequencing was carried out by Sangon Biotech (Shanghai) Co., Ltd. The sequencing results were amplified using universal primers 27 F (AGAGTTTGATCCTGGCTCAG) and 1492R (TACGGYTACCTTGTTAYGACTT) from both ends using forward and reverse primers. The obtained sequences were spliced and compared in NCBI (www.ncbi.nlm.nih.gov) and Ezbiocloud (www.ezbiocloud.net) to determine the homology with known sequences, and those with more than 97% sequence similarity were classified as the same species.
(4) Phylogenetic analysis.
The software MEGA 7.0 was used to compare the sequences of the strains with those of similar strains from Ezbiocloud (www.ezbiocloud.net), and the matrix distance was calculated from the sequence data according to the “Kimura two-parameter” method. The neighbor-joining method was used to perform 1000-replicate validation and phylogenetic tree analysis.
High-throughput sequencing
Sample processing
The endophytic bacteria, epiphytic bacteria and bacteria in the surrounding seawater were named AS, ASE and AW, respectively, and each group had three replicates. Each replicate was from one sampling site.
(1) Endophytic bacteria.
An amount of 2.0 g of disinfected S. thunbergii was placed in lyophilization tubes and stored at -80 °C until DNA extraction.
(2) Epiphytic bacteria.
Epiphytic bacteria samples were obtained in accordance with the previous literature (Mathai et al. 2018). Approximately 25 g of each sample of S. thunbergii was transferred to a 250 mL conical flask, followed by the addition of 70 mL of 0.01 M phosphate buffer (pH 7.4), and then the flask was sealed with sterilized film and shaken for 30 min at room temperature in a rotary HZQ-C shaker (HDL, Harbin, China) at 200 r min-1 to obtain a suspension of epiphytic bacteria isolated from the surface of the algae. The suspension of epiphytic bacteria was filtered through a sterile 500-mesh sieve to remove impurities such as sand mixed in the suspension. The bacteria were collected onto sterile 0.22 μm filter membranes (Millipore, Merck KGaA, Darmstadt, Germany) via filtration in a sterile environment using vacuum filtration system (Steriflip, Millipore, USA), and the membranes were stored at -80 °C until DNA extraction.
(3) Bacteria in the surrounding seawater.
The bacteria in the surrounding seawater (500 mL) were collected onto 0.22 μm membranes via vacuum filtration in a sterile environment and stored at -80 °C until being processed for DNA extraction within a week.
High-throughput sequencing and sequence processing
High-throughput sequencing was conducted by Guangzhou Genedenovo Biotechnology Co., Ltd (Guangzhou, China). The genomic DNA was extracted from the samples, and the V3 + V4 regions of 16S rDNA were amplified with specific primers 341 F (CCTACGGGNGGCWGCAG) and 806R (GGACTACHVGGGTATCTAAT). The PCR amplification products were recovered via gel splicing and quantified using a Quanti Fluor TM fluorometer (Promega, United States). Equal amounts of purified amplification products were mixed and sequencing adapters were attached; then, sequencing libraries were constructed using an Illumina Hiseq2500 PE250 (Illumina, Inc., San Diego, CA, USA).
Data analysis
Sequences were spliced and de-duplicated using Uparse software (v9.2.64_i86linux32) (Edgar 2013), and species with > 97% similarity were clustered into operational taxonomic units (OTU). Chloroplast and mitochondrial sequences were removed from the OTU tables. The obtained OTUs were classified based on the Greengenes database (version gg_13_5, greengenes.secondgenome.com/) and Bacterial SILVA database (SILVA version 106, http://www.arb-silva.de/documentation/release-106/) databases (Janssen et al. 2018), and the pre-processing of data and the selection of OTUs from the amplicons were performed using Mothur pipelines (version 1.39.1 available online: https://mothur.org/wiki/miseq_sop/mothur) (Schloss et al. 2009) to calculate the diversity indices, namely Shannon, Simpson, Ace, the Chao1 index and coverage; β-diversity was calculated and analyzed via unweighted UniFrac distances for principal coordinate analysis (PCoA) and the unweighted pair group method with arithmetic mean (UPGMA). The resulting high-throughput sequencing data were clustered according to OTU classification for species diversity at the phylum and genus levels. Species without a clear classification and species with a relative abundance of less than 1% were jointly recorded as Other, and histograms were drawn. Linear discriminant analysis effect size (LEfSe) software (v1.0, https://huttenhower.sph.harvard.edu/galaxy/) was used to analyze the differences between groups (Qin et al. 2020). In LEfSe, the Kruskal–Wallis rank sum test was first performed among all groups of samples, and the screened differences were then compared between two groups using the Wilcoxon rank sum test. The results were ranked using linear discriminant analysis (LDA) to obtain the LDA variance analysis graph, and then the evolutionary branching graph was obtained by mapping the differences to a classification tree with a known hierarchical structure.
Results
Isolation and identification of culturable bacteria
A total of 54 strains of endophytic bacteria were isolated from S. thunbergii, but only 39 strains were finally identified because some strains preserved in sterilized 0.9% physiological saline containing 15% glycerol at -80℃ were not successfully activated. A phylogenetic evolutionary tree based on the molecular biological identification of the strains is shown in Fig. 1. These strains belong to 2 phyla, 5 genera and 16 species (Table 1). At the phylum level, Firmicutes was overwhelmingly dominant (85.00%), including the three genera Bacillus, Alkalihalobacillus and Metabacillus; the other phylum was Proteobacteria (15.00%), including the two genera Ruegeria and Roseibium. At the genus level, the dominant genera were Bacillus (31.00%), Alkalihalobacillus (28.30%) and Metabacillus (25.70%); at the species level, the dominant species were M. indicus (25.70%), A. hwajinpoensis (15.40%) and B. safensis subsp. safensis (10.30%).
Results of high-throughput sequencing
Biodiversity
α-Diversity
The results of the high-throughput sequencing of the endophytic and epiphytic bacteria of S. thunbergii and bacteria in the surrounding seawater samples were compared and analyzed, and a total of 1,062,077 sequences were obtained. A total of 1,011,972 optimized sequences were obtained after quality filtering and removal of chimera, chloroplast and mitochondrion sequences, and the coverage of each sample was above 99%, as shown in Fig. 2, indicating that the sequencing depth covered most of the bacteria in the samples, and that the sequencing data are reliable and appropriate.
The analysis results of the Chao1 index and ACE index (Table 2) showed that the richness of the endophytic bacteria was in the middle—lower than that of the epiphytic bacteria and higher than that of the seawater samples—but the Shannon index and Simpson index showed that the diversity of the endophytic bacteria was lower than that of the epiphytic bacteria and bacteria in the surrounding seawater. It is worth mentioning that the four diversity indices of the epiphytic bacterial community were higher than those of the other two groups of samples.
β-Diversity
Figure 3 shows the results of the Bray-based UPGMA and PCoA analysis. The clustering analysis showed that the compositions of the endophytic and epiphytic bacterial communities of S. thunbergii and bacterial community in the seawater samples were similar within the groups, and only one sample of endophytic bacteria differed obviously from the other two samples in the group, being similar to the bacterial samples from the seawater. The results of the PCoA analysis showed that the structure of the bacterial communities differed significantly between groups (P < 0.05, ANOSIM test), indicating that the endophytic bacterial communities were different from the bacterial communities of the epiphytic bacteria and seawater samples.
Composition of the bacterial community
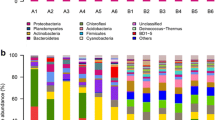
The relative abundance of the endophytic bacterial community structure of S. thunbergii at the phylum and genus levels is shown in Fig. 4A. At the phylum level, the most dominant phylum of the endophytic bacteria was Proteobacteria (42.47%), followed by Cyanobacteria (36.41%), Bacteroides (8.16%), Planctomycetes (4.03%) and Actinomycetes (2.84%). The dominant phyla of the epiphytic bacteria were similar to those of the endophytic bacteria and bacteria in the seawater samples, with the most dominant phylum being Proteobacteria, but its abundance was lower than that in the other two groups of samples (50.15% and 52.37%, respectively). Similarly, Bacteroides, as the third most dominant phylum of the endophytic bacteria, was the second most dominant phylum in the epiphytic bacteria and seawater samples, with abundances of 28.68% and 12.95%, respectively, both of which were higher than the abundance in the endophytic bacteria. However, the abundance of Cyanobacteria in the endophytic bacteria (36.41%) was significantly higher than that in the epiphytic bacteria (9.68%) and seawater samples (2.34%). The abundances of Actinomycetes and Firmicutes in the endophytic bacteria were 2.84% and 1.59%, respectively—in between those of the epiphytic bacteria (1.27%/0.11%) and seawater samples (9.23%/5.97%).
At the genus level (Fig. 4B), the most dominant genus of the endophytic bacteria was Ralstonia (7.53%), followed by Sphingomonas (4.36%), Acaryochloris_MBIC11017 (3.65%). The dominant genera in the epiphytic bacteria and seawater samples were completely different, with the most dominant genus of the epiphytic bacteria being Costertonia (5.58%), followed by Marinomonas (4.47%), Vibrio (3.02%), Acaryochloris_MBIC11017 (1.71%) and Sphingomonas (1.21%), while the most dominant bacteria in the seawater samples were Pseudoalteromonas (6.68%), followed by Candidatus_Actinomarina, (5.07%), Mycoplasma (3.05%), Glaciecola (2.49%) and Vibrio (1.65%).
As can be seen in the Venn diagram (Fig. 5A), 27 phyla were shared by the endophytic bacteria, epiphytic bacteria and seawater samples. There was no specific phylum in the endophytic bacteria, but the specific phyla in the epiphytic bacteria were Cloacimonetes and Rokubacteria; the specific phyla in the seawater samples were Latescibacteria and Synergistetes, indicating that the richness of the endophytic bacteria at the phylum level was lower than that of the epiphytic bacteria and seawater samples.
At the genus level (Fig. 5B), 179 genera were shared by the endophytic bacteria, epiphytic bacteria and seawater samples. Among the shared bacteria, the highest abundance among the endophytic bacteria was found for Acaryochloris_MBIC11017, Sphingomonas and Sphingobium, and the highest abundance among the epiphytic bacteria was found for Costertonia, Marinomonas and Vibrio, while the most abundant bacteria in the seawater samples were Pseudoalteromonas, Candidatus_Actinomarina and Glaciecola. In addition, there were 82 specific genera in the endophytic bacteria, mainly including Hydrogenophaga, Mesorhizobium, Methyloversatilis, Weissella, Fimbriiglobus, Nevskia and Pseudoxanthomona, Arsenophonus, Buchnera; 40 specific genera in the epiphytic bacteria, mainly including Epixenosomes_of_Euplotidium_arenarium, Cupriavidus and Amphiplicatus; and 96 specific genera in the seawater samples, mainly including Capnocytophaga, Providencia, Rothia and Fusobacterium.
Indicator species
LEfSe was used to identify bacterial taxa with significant differences in abundance between groups, i.e. indicator species (Dufrêne and Legendre, 1997) and the results (Fig. 6) showed that the indicator species of endophytes were not the same as those of the epiphytic bacteria and seawater samples. The indicator taxa of the endophytic bacteria were mainly Ralstonia (genus), Hydrogenophaga (genus) and Mesorhizobium (genus), while Bacteroidetes (phylum), Bacteroidia (class) and Flavobacteriales (order) were enriched in the epiphytic bacteria, and Mollicutes (class), Fusobacteria (phylum) and Mycoplasmatales (order) were abundant in the seawater samples.
Figure 7 showed the bacteria with significant differences in abundance at phylum level between endophytic, epiphytic bacteria of S. thunbergii and seawater samples at the phylum level (P < 0.05). Only Planctomycetes had the largest indicator value in endophytic bacteria while Bacteroidetes, Verrucomicrobia and Deinococcus-Thermus were enriched in epiphytic bacteria. On the contrary, these phyla with high indicator values in endophytic or epiphytic bacteria had low indicator values in seawater samples, while Actinobacteria, Fusobacteria, Tenericutes and Epsilonbacteraeota with low abundance in endophytic or epiphytic bacteria were enriched in seawater samples.
Discussion
In this study, the community structure of the endophytic bacteria in S. thunbergii was investigated using a combination of the traditional culture method and high-throughput sequencing technology, the bacterial community structure of the epiphytic bacteria and bacteria in seawater samples was also analyzed. The results showed that the endophytic bacterial communities of S. thunbergii had some dominant phyla in common with those of other marine macroalgae and terrestrial plants, but the dominant genera were very different.
The dominant phyla of the endophytic bacteria of S. thunbergii obtained using the culture method were Firmicutes and Proteobacteria and these two phyla were also dominant in K. alvarezii (Mangun et al. 2023). This result was also consistent with the previous studies showing that the dominant phylum of the endophytic bacteria of terrestrial plants was Proteobacteria, followed by a small number of Firmicutes, Actinomycetes and Bacteroides (Trivedi et al. 2020; Edwards et al. 2015). However, the dominant genus and species of the endophytic bacteria in S. thunbergii were different from those in K. alvarezii (Mangun et al. 2023). Alkalihalobacillus, Metabacillus, Ruegeria and Roseibium were reported as plant endophytes for the first time, but these bacteria are common in marine environments or marine organisms. For example, Alkalihalobacillus is a salt-tolerant bacterium (Li et al. 2021); Metabacillus has been reported to induce coral larval sedimentation and metamorphosis (Zhang et al. 2021b); Ruegeria is common in cultured waters, having been reported to be present in the intestine of Anguilla rostrata (Liu et al. 2021a); Roseibium spp. are the dominant intestinal species in healthy Hippocampus (Yan et al. 2020). At the species level, the endophytic nitrogen-fixing bacterium M. indicus was the most abundant, indicating that endophytic bacteria may play a certain role in the acquisition of nitrogen in algae, followed by A. hwajinpoensis, which is a salt-tolerant strain with a denitrification function (Cao et al. 2019). The third most dominant bacterium was B. safensis subsp. safensis, which has been reported many times in terrestrial plants and is a biocontrol endophyte with a wide range of antibacterial activities against a variety of pathogenic fungi and bacteria (Zhang et al. 2020b).
The results of the high-throughput sequencing revealed that the dominant phyla of the endophytic bacteria of S. thunbergii were Proteobacteria, Cyanobacteria, Bacteroides, Planctomycetes and Actinomycetes This result was similar to that of the dominant phylum of endophytic bacteria in S. horneri and U. prolifera (Mei 2019), but it varied considerably at the genus level, without the same dominant genera. The most dominant genus of the endophytic bacteria was Ralstonia. Ralstonia solanacearum, the pathogen of a devastating worldwide crop bacterial disease, is a member of this genus (She and He 2020). Other dominant genera include important degraders in the environment (Liu et al. 2017), such as Sphingomonas, Hydrogenophaga and Sphingobiumand Cyanobacteria, Acaryochloris_MBIC11017. Further analysis of other genera of endophytic bacteria revealed that the most abundant taxa were bacteria with degradation functions, including the highly efficient benazolin-ethyl degrading Methyloversatilis (Qian et al. 2011), chitinolytic Fimbriiglobus (Ravin et al. 2018), pyrene-degrading Pseudoxanthomonas (Klankeo et al. 2009), cellulolytic Ferrovibrio (Dahal et al., 2018), denitrifying Janthinobacterium (Yang 2019), organic-matter-degrading Lysinibacillus (Li et al. 2018; Zhu 2015), members of Pseudorhodoplanes (Zhen et al. 2019), which degrade complex organic matter, and members of Phenylobacterium, comprising the best indigenous petroleum-hydrocarbon-degrading bacteria, suggesting that endophytic bacteria have various strong degradation functions. In addition, there were also multiple nitrogen-fixing bacteria, such as Mesorhizobium (Yang 2017) and some members of Cyanobacteria, including Arthrospira_PCC-7345 (Du et al. 2016), Trichodesmium_IMS101 (Kranz et al. 2010), Rhodobacter (Jin et al. 2019) and Niveispirillum (Cai et al. 2018). There were also some bacteria with antibacterial activity, or bacteria that are generally considered beneficial, such as the new antibacterial agent and probiotic Weissella, (Tenea and Hurtado 2021), Arsenophonus which contributes to the resistance of Nilqparvata lugens (Chen et al. 2014), Buchnera (Li and Li 2006), whose members are specialized symbiotic bacteria that provide a variety of essential amino acids and B vitamins to the host aphid, and intestinal beneficial bacteria belonging to Bifidobacterium (Li and Zhu 2022). It was interesting that there are also some pathogenic bacteria among the endophytic bacteria, including the most dominant genus Ralstonia comprises pathogens that causes plant bacterial wilt and fish pathogen Plesiomonas (Chen et al. 2020), Neorickettsia, Legionella and Acidovorax (Zhang et al. 2019). In summary, the composition and function of the endophytic bacterial community of S. thunbergii are very complicated. How these bacteria interact with the host S. thunbergii and finally form a stable symbiosis within S. thunbergii needs to be further investigated.
In addition, the community composition of the endophytic bacteria of S. thunbergii were significantly different compared to those of the epiphytic bacteria and bacteria in the seawater samples. Although 27 phyla and 179 genera were shared, the 3 groups of samples could be clustered separately in the β-diversity analysis, indicating the variability between the bacterial communities from the three types of samples. Firstly, the relative abundance of the dominant phyla differed significantly. The abundance of Cyanobacteria was absolutely dominant, far exceeding the abundance in the epiphytic bacteria and in the seawater samples, indicating that the symbiotic cyanobacteria of S. thunbergii mainly live inside the algal body. The abundance at the genus level also varied greatly, and some genera were highly abundant inside the algae, such as Acaryochloris_MBIC11017, which was obviously higher in the endophytic bacteria than in the epiphytic bacteria and seawater samples; this was consistent with the result that the phylum Cyanobacteria was dominant in the endophytic bacteria, indicating that a large number of Cyanobacteria live inside of S. thunbergii. There were also some bacteria with a low abundance in the endophytic bacteria, such as the marine oil-degrading Pseudoalteromon (Zhang et al. 2020a), with an obviously lower abundance in the endophytic bacteria than in the epiphytic bacteria and the seawater samples. This suggests that the taxa of endophytic bacteria and their functions differ significantly from those of bacteria on the surface of macroalgae and in the surrounding seawater. In addition, some pathogenic bacteria were also found in extremely low abundance inside the seaweed, such as Mycoplasma, whose abundance inside the macroalgae was much lower than in the epiphytic bacteria and seawater samples. This was also the case for Vibrio. The low abundance of these pathogen in the endophytic bacteria of S. thunbergia suggests that the cell wall of S. thunbergii is resistant to the invasion of pathogenic bacteria.
The endophytic bacteria also differed from the epiphytic bacteria and bacterial communities in the seawater samples in the indicator species and specific bacteria of each group. For example, the indicator species Ralstonia was highly abundant in endophytic communities; however, it was not found in the epiphytic bacteria, and it only had an abundance of 0.02% in the seawater samples. Ralstonia, includes Ralstonia solanacearum, an important pathogen that causes wilt disease in many land plants (She and He 2020), as well as many strains of organic pollutants-degrading bacteria (Coll et al. 2020) and phosphate-solubilizing (Dandessa and Bacha 2018). Ralstonia was also found in plant seeds (Liu et al. 2011), indicating that Ralstonia can be transmitted from parents to offspring without the need to enter the plant body from the external environment. This may be the reason why Ralstonia was not present in the epiphytic bacteria in S. thunbergii in this study, while its abundance was high in the endophytic bacteria.
Similarly, the degrading bacterium Hydrogenophaga (Liu et al. 2017) was enriched in the endophytic bacteria, but this genus was absent in both the epiphytic bacteria and bacteria in the seawater samples, suggesting that not all endophytic bacteria in S. thunbergii enter from the surrounding seawater. In contrast, the human pathogenic bacterium Capnocytophaga (Ye et al. 2018) was enriched in the seawater samples, but it was absent in both the endophytic and epiphytic bacteria, suggesting that it selectively enters S. thunbergii and forms a community different from that in the surrounding seawater and on the algal surface.
Both the culture method and high-throughput sequencing technology indicated that there were abundant endophytic bacterial groups in S. thunbergii. Moreover, the number of bacterial taxa obtained using high-throughput sequencing technology was much higher than that obtained using the culture method, and the dominant bacterial taxa obtained using these two methods were different. This may be mainly due to the limitations of the culture method, where only approximately 1% of the bacteria found in the natural environment are cultivable (Eilers et al. 2000). Endophytic bacteria inhabit the interiors of plants, whose community composition is closely related to the internal physiological and biochemical conditions of host plants. Due to the fact that during in vitro cultivation, the bacterial culture medium does not contain algae-derived substances, many groups of bacteria cannot be cultivated (Mitrani et al. 2005). Further, the commonly used method of isolating endophytic bacteria from plants actually isolates aerobic bacteria. Although the algal cell walls are permeable, the results of the high-throughput sequencing in this study revealed that the endophytic bacteria include anaerobic bacteria such as Hydrogenophaga (Tan et al. 2021) and Nevskia (Li 2017), which cannot grow under aerobic conditions. Therefore, the taxa of the endophytic bacteria isolated using the culture method in this study were fewer in number than those obtained using high-throughput sequencing, but the culture method can isolate and obtain a large number of purified strains for further studies of bacteria–algae interactions, as well as the development of functional strains. Therefore, it is still important in the study of endophytic bacteria in algae.
In conclusion, the composition of the endophytic bacterial community of S. thunbergii was studied using the culture method and high-throughput sequencing technology in this study. It was found that the endophytic bacteria in S. thunbergii are abundant and enriched in groups with nitrogen fixation, salt tolerance, pollutant degradation, and antibacterial properties as well as pathogenic bacteria. In addition, the communities of the endophytic bacteria differed from that of epiphytic bacteria in S. thunbergii and the bacteria in the surrounding seawater. This study provides a preliminary understanding of the structure of the endophytic bacterial community of S. thunbergii and helps to elucidate the mechanism of bacterial–algal relationships. Moreover, a large number of endophytic bacterial strains were obtained, providing experimental materials for the effective utilization and development of bacterial resources.
Data availability
All bacterial 16S rRNA gene sequence data produced during the study were deposited in the NCBI Sequence Read Archive database under Bioproject PRJNA835680. Results for concurrent bacterial cultures are available in the NCBI SRA repository under the BioProject ID: PRJNA841409.
References
Ahmed EF, Hassan HM, Rateb ME, Abdel-Wahab N, Sameer S, Ebel R, Taie HAA, Hameed MSA, Hammouda O (2016) A comparative biochemical study on two marine endophytes, Bacterium SRCnm and Bacillus Sp. JS, Isolated from red sea algae. Pak J Pharm Sci. 29:17–26
Benjelloun I, Thami Alami I, Douira A, Udupa SM (2019) Phenotypic and genotypic diversity among symbiotic and non-symbiotic bacteria present in Chickpea nodules in Morocco. Front Microbiol 10:1885. https://doi.org/10.3389/fmicb.2019.01885
Cai H, Cui H, Zeng Y, Wang Y, Jiang H (2018) Niveispirillum lacus sp. nov., isolated from cyanobacterial aggregates in a eutrophic lake. Int J Syst Evol Microbiol 68:507–512. https://doi.org/10.1099/ijsem.0.002526
Cai Y, Liu H, Wang Y, Xie J, Huang J, Lao J, He W, Zhang S (2020) Analysis of structure and diversity of endophytic bacteria community in Polygonatum cyrtonema and diversity based on 16S rRNA gene high-throughput sequencing. J Hunan University of Chinese Medicine. 40:846–852. https://doi.org/10.3969/j.issn.1674-070X.2020.07.013
Cao K, Deng G, Hu X, Zhang J, Wu C (2019) Screening of a halotolerant Bacillus and its enzymological properties of protease. J Anhui Agricultural Univ 46:312–317. https://doi.org/10.13610/j.cnki.1672-352x.20190320.018
Chen Y, Chen Y, Wang W, Lai F, Fu Q (2014) A preliminary study on the transfer mode and biological significance of endosymbiont Arsenophonus in Nilaparvata lugens. Chin J Rice Sci 28:92–96. https://doi.org/10.3969/j.issn.1001-7216.2014.01.013
Chen M, Wang D, Pan Y, Zhang C, Zha XLM, Tian Y (2020) A review of research progress on pathogenic Plesiomonas shigelloides from fish. Fisheries Sci 39:780–786. https://doi.org/10.16378/j.cnki.1003-1111.2020.05.018
Coll C, Bier R, Li Z, Langenheder S, Gorokhova E, Sobek A (2020) Association between aquatic micropollutant dissipation and river sediment bacterial communities. Environ Sci Technol 54:14380–14392. https://doi.org/10.1021/acs.est.0c04393
Dahal RH, Kim J (2018) Ferrovibrio soli sp. nov., a novel cellulolytic bacterium isolated from stream bank soil. Int J Syst Evol Microbiol 68:427–431. https://doi.org/10.1099/ijsem.0.002527
Dandessa C, Bacha K (2018) Review on role of phosphate solubilizing microorganisms in sustainable agriculture. Int J Curr Res Aca Rev 6:48–55. https://doi.org/10.20546/ijcrar.2018.611.006
de Jong H, Reglinski T, Elmer PAG, Wurms K, Vanneste JL, Guo LF, Alavi M (2019) Integrated use of Aureobasidium pullulans strain CG163 and Acibenzolar-S-Methyl for management of bacterial canker in kiwifruit. Plants (Basel) 8:287. https://doi.org/10.3390/plants8080287
Du G, Wang F, Hu T, Wang L, Chen W (2016) Characterization of phospholipid fatty acid of two Arthrospira strains. J Huazhong Agricultural Univ 35:11–16. https://doi.org/10.13300/j.cnki.hnlkxb.2016.06.002
Dufrêne M (1997) P. Legendre Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67 345–366 https://doi.org/10.1890/0012-9615(1997)067[0345,SAAIST]2.0.CO;2
Edgar R (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112:E911–E920. https://doi.org/10.1073/pnas.1414592112
Eilers H, Pernthaler J, Glöckner FO, Amann R (2000) Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. https://doi.org/10.1128/AEM.66.7.3044-3051.2000. 66:3044 ~ 3051
Feng S, Xie G, Liu N, Xu C, Lu Y, Feng X (2020) Isolation and antioxidative activities of algal endophytes. J Food Sci Biotechnol 39:99–105. https://doi.org/10.3969/j.issn.1673-1689.2020.06.014
Guo Y, Qin Y, Ju T, Leng S, Wu D, Li M (2016) Isolation and characterization of a strain endophytic bacterium in Myriophyllum spicatum and its solubilization of phosphate. Acta Sci Circum 36:4352–4360. https://doi.org/10.13671/j.hjkxxb.2016.0174
Hollants J, Leroux O, Leliaert F, Decleyre H, De Clerck O, Willems A (2011) Who is in there? Exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta). PLoS ONE 6:e26458. https://doi.org/10.1371/journal.pone.0026458
Janssen S, Mcdonald D, Gonzalez A, Navas-molina JA, Jiang L, Xu Z (2018) Phylogenetic placement of exact amplicon squences improves associations with clinical information. MSystems. 3: e00021-00018. https://doi.org/10.1128/mSystems.00021-18
Jin X, Hu W, He S, Zhou T, Wang Y, Zhong Z (2019) Diversity of soil nitrogen-fixing microorganisms in Salicornia europaea community of Ebinur Lake Wetland during different periods. Acta Microbiol Sinica 59:1600–1611. https://doi.org/10.13343/j.cnki.wsxb.20180591
Klankeo P, Nopcharoenkul W, Pinyakong O (2009) Two novel pyrene-degrading Diaphorobacter sp. and Pseudoxanthomonas sp. isolated from soil. J Bioscience Bioeng 108:488–495. https://doi.org/10.1016/j.jbiosc.2009.05.016
Kranz SA, Levitan O, Richter KU, Prásil O, Berman-Frank I, Rost B (2010) Combined effects of CO 2 and light on the N2 -fixing cyanobacterium Trichodesmium IMS101: physiological responses. Plant Physiol 154:334–345. https://doi.org/10.1104/pp.110.159145
Kunda P, Dhal PK, Mukherjee A (2018) Endophytic bacterial community of rice (Oryza sativa L.) from coastal saline zone of West Bengal: 16S rRNA gene based metagenomics approach. Meta Gene 18:79–86. https://doi.org/10.1016/j.mgene.2018.08.004
Lan J, Zhu Y, Su M, Ge C, Liu Y, Liu B (2008) Isolation and identification of the endophyte bacteria from Eichhornia crassipe (Mart.) Solms. J Agro-Environment Sci 27:2423–2429. https://doi.org/10.3321/j.issn:1672-2043.2008.06.053
Li T (2017) Isolation of anaerobe from wastewater and sea sediment and analysis of strain YJ1. MSc Dissertation. Zhejiang University
Li X, Li B (2006) Interactions among aphids and their endosymbiotic Buchnera and other secondary symbionts. Chin Bull Entomol 43:443–447. https://doi.org/10.3969/j.issn.0452-8255.2006.04.003
Li H, Zhu S (2022) Recent advances in the role and mechanism of Bifidobacterium in major human diseases. Chin J Anim Infect Dis 30:206–214. https://doi.org/10.19958/j.cnki.cn31-2031/s.20220412.001
Li J, Zhou W, Li F, Gao C, Yi X (2017) Diversity of cultivated marine bacteria and antibacterial activity of endophytic bacterial in Rhizophora Stylosa. Guihaia 37:308–314. https://doi.org/10.11931/guihaia.gxzw201604019
Li G, Zeng X, Zhai L, Leng Y, Liu M, Li S, Chen T (2018) Screening, identification and characteristics of Lysinibacillus fusiformis 23 – 1 for petroleum degradation. Acta Agriculturae Zhejiangensis 30:1229–1236. https://doi.org/10.3969/j.issn.1004-1524.2018.07.17
Li Y, Gao T, Dong J, Zhou T, Yu Y (2021) Diversity of endophytic bacteria in Spirodela polyrrhiza analyzed by high-throughput sequencing technology. J Hunan Ecol Sci 8:54–60. https://doi.org/10.3969/j.issn.2095-7300.2021.03.009
Liu Y, Zuo S, Zou YY, Wang JH, Song W (2011) Diversity of endophytic bacterial communities in seeds of hybrid maize (Zea mays L., Nongda108) and their parental lines. Scientia Agricultura Sinica 44:4763–4771. https://doi.org/10.3864/j.issn.0578-1752.2011.23.001
Liu X, Wang Y, Xu H, Jiang H, Jiang Y, Song N (2017) Effects of microbial communities on the composition of algae-derived and grass-derived chromophoric dissolved organic matter in Eutrophic Lake. Ecol Environ Sci 26:1403–1409. https://doi.org/10.16258/j.cnki.1674-5906.2017.08.016
Liu H, Liang Y, Zhai S (2021a) Effect of different starter feeds on intestinal flora of Anguilla rostrata at elver stage. Feed Res 44:51–55. https://doi.org/10.13557/j.cnki.issn1002-2813.2021.05.013
Liu L, Ming X, Zhang X, Hao J, Fu L, Wang Q, Lyu X, Chen W, Liu Q (2021b) Diversity of endophytic bacteria in faba bean seeds by high-throughput sequencing. J Agricultural Sci Technol 23:73–80. https://doi.org/10.13304/j.nykjdb.2019.0860
Liu LH, Yuan T, Zhang JY, Tang GX, Lü H, Zhao HM, Li H, Li YW, Mo CH, Tan ZY, Cai QY (2022) Diversity of endophytic bacteria in wild rice (Oryza Meridionalis) and potential for promoting plant growth and degrading phthalates. Sci Total Environ 806:150310. https://doi.org/10.1016/j.scitotenv.2021.150310
Lu C, Wang Q, Jiang Y, Zhang M, Meng X, Li Y, Liu B, Yin Z, Liu H, Peng C, Li F, Yue Y, Hao M, Sui Y, Wang L, Cheng G, Liu J, Chu Z, Zhu C, Dong H, Ding X (2022) Discovery of a novel nucleoside immune signaling molecule 2’-deoxyguanosine in microbes and plants. J Adv Res 46:1–15. https://doi.org/10.1016/j.jare.2022.06.014
Mancuso FP, D’Hondt S, Willems A, Airoldi L, De Clerck O (2016) Diversity and temporal dynamics of the epiphytic bacterial communities associated with the canopy-forming seaweed Cystoseira compressa (Esper) Gerloff and Nizamuddin. Front Microbiol 7:476. https://doi.org/10.3389/fmicb.2016.00476
Mangun VV, Sugumaran R, Lym Yong WT, Yusof NA (2023) Dataset of 16S ribosomal DNA sequence-based identification of endophytic bacteria isolated from healthy and diseased Sabah red algae, Kappaphycus Alvarezii. Data Brief 51:109785. https://doi.org/10.1016/j.dib.2023.109785
Mathai PP, Dunn HM, Magnone P, Brown CM, Chun CL, Sadowsky MJ (2018) Spatial and temporal characterization of epiphytic microbial communities associated with eurasian watermilfoil: a highly invasive macrophyte in North America. FEMS Microbiol Ecol 94:12–21. https://doi.org/10.1093/femsec/fiy178
Mei X (2019) Community structures and functions of bacteria associated with blooming seaweeds in the Yellow Sea. MSc Dissertation. University of Chinese Academy of Sciences
Melo R, Sousa-Pinto I, Antunes SC, Costa I, Borges D (2021) Temporal and spatial variation of seaweed biomass and assemblages in Northwest Portugal. J SEA RES 174:102079. https://doi.org/10.1016/j.seares.2021.102079
Mitrani E, Nadel G, Hasson E, Harari E, Shimoni Y (2005) Epithelial-mesenchymal interactions allow for epidermal cells to display an in vivo-like phenotype in vitro. Differentiation 73:79–87. https://doi.org/10.1111/j.1432-0436.2005.07302002.x
Pei P, Aslam M, Du H, Liang H, Wang H, Liu X, Chen W (2021) Environmental factors shape the epiphytic bacterial communities of Gracilariopsis Lemaneiformis. Sci Rep 11:8671. https://doi.org/10.1038/s41598-021-87977-3
Peng LH, Liang X, Xu JK, Dobretsov S, Yang JL (2020) Monospecific biofilms of Pseudoalteromonas promote larval settlement and metamorphosis of Mytilus coruscus. Sci Rep 10:2577. https://doi.org/10.1038/s41598-020-59506-1
Puri RR, Adachi F, Omichi M, Saeki Y, Yamamoto A, Hayashi S, Ali MA, Itoh K (2019) Metagenomic study of endophytic bacterial community of sweet potato (Ipomoea batatas) cultivated in different soil and climatic conditions. World J Microbiol Biotechnol 35:176. https://doi.org/10.1007/s11274-019-2754-2
Purushotham N, Jones E, Monk J, Ridgway H (2020) Community structure, diversity and potential of endophytic bacteria in the primitive New Zealand medicinal plant pseudowintera colorata. Plants (Basel) 9:156. https://doi.org/10.3390/plants9020156
Qian L, Chen L, Ren Q, Gu H, Cai T (2011) Isolation and identification of a benazolin-ethyl degrading strain and its degradation characteristics. Eenvironmental Sci 32:1805–1811. https://doi.org/10.13227/j.hjkx.2011.06.010
Qin W, Song P, Lin G, Huang Y, Wang L, Zhou X, Li S, Zhang T (2020) Gut microbiota plasticity influences the adaptability of wild and domestic animals in co-inhabited areas. Front Microbiol 11:125. https://doi.org/10.3389/fmicb.2020.00125
Ran H, Kong W, Jiang H, Shi Y, Xin Z (2016) Isolation and identification of endophytic bacteria from Salicornia bigelovii torr. And their antimicrobial activities. Food Ferment Industries 42:79–86. https://doi.org/10.13995/j.cnki.11-1802/ts.201603014
Ravin NV, Rakitin AL, Ivanova AA, Beletsky AV, Kulichevskaya IS, Mardanov AV, Dedysh SN (2018) Genome analysis of fimbriiglobus ruber SP5 T, a planctomycete with confirmed chitinolytic capability. Appl Environ Microbiol 84:e02645–e02617. https://doi.org/10.1128/AEM.02645-17
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
She X, He Z (2020) Advances in studies on crop bacterial wilt caused by ralstonia solanacearum. Guangdong Agricultural Sci 47:82–89. https://doi.org/10.16768/j.issn.1004-874x.2020.12.009
Singh RP, Baghel RS, Reddy CR, Jha B (2015) Effect of quorum sensing signals produced by seaweed-associated bacteria on carpospore liberation from Gracilaria dura. Front Plant Sci 6:117. https://doi.org/10.3389/fpls.2015.00117
Subramanian M, Marudhamuthu M (2020) Hitherto unknown terpene synthase organization in taxol-producing endophytic bacteria isolated from marine macroalgae. Curr Microbiol 77:918–923. https://doi.org/10.1007/s00284-020-01878-8
Sugrani A, Ahmad A, Djide MN, Natsir H (2019) Toxicological evaluation and antibacterial activity of crude protein extract from endophytic bacteria associated with algae Eucheuma Spinosum. J Phys Conf Ser 1341:032006. https://doi.org/10.1088/1742-6596/1341/3/032006
Tan X, Yang YL, Liu YW, Yin WC, Fan XY (2021) The synergy of porous substrates and functional genera for efficient nutrients removal at low temperature in a pilot-scale two-stage tidal flow constructed wetland. Bioresour Technol 319:124135. https://doi.org/10.1016/j.biortech.2020.124135
Tenea GN, Hurtado P (2021) Next-generation sequencing for whole-genome characterization of Weissella cibaria UTNGt21O strain originated from wild Solanum quitoense lam. Fruits: an atlas of metabolites with biotechnological significance. Front Microbiol 12:675002. https://doi.org/10.3389/fmicb.2021.675002
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18:607–621. https://doi.org/10.1038/s41579-020-0412-1
Wang Z, Ji Y, Chen Y (2015) Studies and biological significances of plant endophytes. Microbiol China 42:349–363. https://doi.org/10.13344/j.microbiol.china.130815
Wang J, Yang Z, Wang G, Shang S, Tang X, Xiao H (2022) Diversity of epiphytic bacterial communities on male and female Sargassum thunbergii AMB Express. 12:97. https://doi.org/10.1186/s13568-022-01439-1
Wu H, Liu H, Zhan D, Li M (2010) Research present situation of Sargassum Thunbergii. Nat Resour Study 1:95–96. https://doi.org/10.3969/j.issn.1003-7853.2010.01.048. Territory
Yan Y, Chen M, Wu R, Wu S, Luo H, Chen X, He L, Zheng L, Huang Z (2020) Analysis on the structural characteristics of intestinal bacteria in Hippocampus infected with gastroenteritis. Fujian Agricultural Sci Technol 1–8. https://doi.org/10.13651/j.cnki.fjnykj.2020.12.001
Yang F (2017) Soil profiles microbial community responses to environmental disturbance and growth of legumes. MSc Dissertation, Northwest A&F University
Yang M (2019) Research on heterotrophic nitrification-aerobic denitrification characterisation and psychrotrophic mechanism of psychrotrophic strain Janthinobacterium sp. M-11. PhD Dissertation, Harbin Institute of Technology
Ye J, Zhang J, Zhang X, Luo B, Chen H (2018) Early onset neonatal septicemia caused by Capnocytophaga sputigena: one case report. Chin J Infect Chemother 18:328–330. https://doi.org/10.16718/j.1009-7708.2018.03.018
Zhang K (2017) Screening manganese resistant endophytes from Myriophyllum and evaluating their potential application in phytobial remediation. MSc Dissertation, Guangxi University
Zhang Y, Han Z, Sun J, Lyu A, Hu X, Liu J (2019) Effects of infection with Shewanella algae on the microbial communities and expression of related functional genes in the intestine of Cynoglossus semilaevis. J South Agric 50:2300–2307. https://doi.org/10.3969/j.issn.2095-1191.2019.10.21
Zhang A, Cheng P, Tang N, Cao J (2020a) Optimizing fermentation condition for petroleum degradation by Pseudoalteromon SI-JHS. Shandong Chem Ind 49:70–73. https://doi.org/10.19319/j.cnki.issn.1008-021x.2020.05.024
Zhang Z, Hu L, Liu L, Ji M (2020b) Identification and antibacterial properties of an antagonistic bacterium Bacillus safensis against walnut fungal disease. J Henan Agricultural Sci 49:97–104. https://doi.org/10.15933/j.cnki.1004-3268.2020.12.014
Zhang Y, Zhang A, Zhang H, Zhang M, Xu Y, Qu S, Miao Y (2021a) Review of endophyte bacteria in Suaeda suaeda(L). Guangxi Plant Prot 34:20–24. https://doi.org/10.3969/j.issn.1003-8779.2021.03.006
Zhang Y, Zhang Y, Yang Q, Ling J, Tang X, Zhang W, Dong J (2021b) Complete genome sequence of Metabacillus Sp. CB07, a bacterium inducing settlement and metamorphosis of coral larvae. Mar Genom 60:100877. https://doi.org/10.1016/j.margen.2021.100877
Zhang S, Sun C, Liu X, Liang Y (2023) Enriching the endophytic bacterial microbiota of Ginkgo roots. Front Microbiol 14:1163488. https://doi.org/10.3389/fmicb.2023.1163488
Zhen L, Wu G, Yang J, Jiang H (2019) Distribution and diversity of sulfur-oxidizing bacteria in the surface sediments of tibetan Hot Springs. Acta Microbiol Sinica 59:1089–1104. https://doi.org/10.13343/j.cnki.wsxb.20190100
Zhu C (2015) Identification of the PBDEs degradation strain Lysinibacillus varians GY32 and the molecular mechanisms of the filament-to-rod cell cycle. PhD Dissertation, South China University of Technology
Acknowledgements
We thank MDPI English Editing (https://www.mdpi.com/authors/english) for editing the English text of a draft of this manuscript. We thank Home for Researchers.
Funding
This work was supported by the National Natural Science Foundation of China (42176154), NSFC-Shandong Joint Fund (U1806213), National Key R&D Program of China (2019YFD0901204).
Author information
Authors and Affiliations
Contributions
Investigation, Data collection, Formal analysis, Writing - original draft & editing, Y.L. and J.W.; Investigation, Data curation, Formal analysis, T.S., X.Y., Z.Y. and Y.Z.; Writing - review & editing, Project administration, Funding acquisition, X.T.; Writing - review & editing, H.X. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Wang, J., Sun, T. et al. Community structure of endophytic bacteria of Sargassum thubergii in the intertidal zone of Qingdao in China. AMB Expr 14, 35 (2024). https://doi.org/10.1186/s13568-024-01688-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-024-01688-2