Abstract
Unlike in chickens, dynamics of the gut microbiome in turkeys is limitedly understood and no data were yet published in context of pathological changes following experimental infection. Thus, the impact of Histomonas meleagridis-associated inflammatory changes in the caecal microbiome, especially the Escherichia coli population and their caecal wall invasion in turkeys was investigated. Birds experimentally inoculated with attenuated and/or virulent H. meleagridis and non-inoculated negative controls were divided based on the severity of macroscopic caecal lesions. The high throughput amplicon sequencing of 16SrRNA showed that the species richness and diversity of microbial community significantly decreased in severely affected caeca. The relative abundances of operational taxonomic units belonging to Anaerotignum lactatifermentans, E. coli, and Faecalibacterium prausnitzii were higher and paralleled with a decreased abundances of those belonging to Alistipes putredinis, Streptococcus alactolyticus, Lactobacillus salivarius and Lactobacillus reuteri in birds with the highest lesion scores. Although the relative abundance of E. coli was higher, the absolute count was not affected by the severity of pathological lesions. Immunohistochemistry showed that E. coli was only present in the luminal content of caecum and did not penetrate even severely inflamed and necrotized caecal wall. Overall, it was demonstrated that the fundamental shift in caecal microbiota of turkeys infected with H. meleagridis was attributed to the pathology induced by the parasite, which only led to relative but not absolute changes in E. coli population. Furthermore, E. coli cells did not show tendency to penetrate the caecal tissue even when the intestinal mucosal barriers were severely compromised.
Similar content being viewed by others
Introduction
A normal gut microbial community and integrity of the intestinal wall in poultry are essential for nutrient digestion, production of beneficial short chain fatty acids and prevention of disease by competitive exclusion or blocking of pathogen colonization [1, 2]. Any shifts of the resident microbiota might compromise all or some of these functions [3] leading to reduction in intestinal barrier function potentially resulting in bacterial translocation from gut to systemic organs [4, 5]. The knowledge on the composition of complex gut microbiome has enhanced with the application of next generation sequencing [6]. Applying this technique, intestinal microbiota and their interactions with the chicken host have been elucidated in experimental infection models with several pathogens, for instance, infection with Eimeria tenella led to relative increase of Enterobacteriaceae and decrease of Lactobacillaceae [7,8,9,10]. Likewise, Histomonas meleagridis infection resulted in reduced gut microbial richness and diversity favouring caecal colonization of avian pathogenic Escherichia coli in chicken layers [4]. In turkeys, such studies are still limited and dynamics of enteric microbiota, especially in the presence of inflammation and necrosis remains to be understood. Previous studies in turkeys focused on characterization of the microbiota along the gastrointestinal tract or decipher changes induced by antibiotic treatment, different litter management or natural infection with haemorrhagic enteritis virus [11,12,13,14]. Additional studies associated composition of turkey gut microbiota with production of short chain fatty acids in the gut or increase in the body weight [15,16,17].
Escherichia coli is a Gram-negative bacterium, a member of the Enterobacteriaceae and resides in the lower digestive tract of chickens and turkeys where it colonizes in the first 24 h post-hatching [18, 19]. Avian colibacillosis caused by E. coli is reported to be responsible for fibrinous peritonitis, hepatitis, polyserositis in turkeys and causing economic losses [20, 21]. Furthermore, some of the E. coli isolates from clinical cases of turkey cellulitis were characterized to be avian pathogenic E. coli based on the presence of virulence associated genes and phylogenetic groups [22]. Although the demarcations between pathotypes are not very clear, some of the intestinal commensals might carry virulence associated genes and the isolates could show opportunistic or potentially pathogenic nature in birds [23]. In contrast to chickens, experimental and field studies related to E. coli infection in turkeys are very scarce. The majority of experimental studies in turkeys involved respiratory routes of infection and were focused to elucidate the impact of stress factors or other co-residing pathogens in the progression of E. coli infection [24,25,26,27]. Thus, the relationship between E. coli populations residing in the gut and systemic bacteremia in turkeys is largely unknown.
Histomonas meleagridis, a protozoal parasite is very well known to cause severe inflammation and necrosis in caeca and liver of experimentally infected turkeys, and in vitro mutual interaction of E. coli with the parasite has been shown previously [28]. In addition, in a previous co-infected model in chicken layers, H. meleagridis enhanced the penetration of E. coli throughout the caecal tissue [4]. The present study was therefore conducted in turkeys with the following two main objectives: (i) to investigate the dynamics of intestinal E. coli population and a possible microbial shift related to the pathology induced by H. meleagridis infection in the gut, and (ii) to assess the consequences of inflammation and necrosis on caecal wall penetration with E. coli and their systemic translocation.
Materials and methods
Birds and housing
Thirty-three one-day-old commercial Hybrid Converter turkeys (Hendrix Genetics, Boxmeer, the Netherlands) were randomly divided into four groups (Table 1). Each group of birds was placed in a separate negatively pressured room on deep litter.
Preparation of H. meleagridis cultures for inoculation
The virulent (in vitro passage of 28 times) or attenuated (in vitro passage of 301 or 302 times) clonal culture of H. meleagridis/Turkey/Austria/2922-C6/04 co-cultivated with the bacterial strain E. coli DH5α, as a supplement for propagation of the parasite [28] was used for inoculation. The inocula medium consisted of Medium 199 with Earle’s salts, l-glutamine, 25 mM HEPES and l-amino acids (Gibco™ Invitrogen, Austria), 15% fetal calf serum (Gibco™ Invitrogen) and rice starch, 0.25% (w/v) (Sigma-Aldrich, Vienna, Austria). The number of viable attenuated and virulent H. meleagridis cells was determined using trypan blue stain and a Neubauer hemocytometer (Sigma-Aldrich, St. Louis, MO, USA) to adjust the appropriate number in each inoculum. For E. coli DH5α, colony-forming units (CFU) were counted from serial dilutions of the inoculum on coliform agar plates after incubation at 37 °C for 24 h.
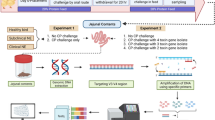
Experimental design
As illustrated in Table 1, different groups of birds were treated as follows: control birds (n = 9) were inoculated with culture media containing only 7.2 × 106 CFU and 1 × 107 CFU of E. coli DH5α at 1 and 28 days of life, respectively; vaccinated birds (n = 9) were inoculated with 6 × 105 cells of attenuated H. meleagridis (passage 301) co-cultured with 1.2 × 108 CFU of E. coli DH5α at 28 days of life; infected birds (n = 6) were inoculated with 6 × 105 cells of virulent H. meleagridis (passage 28) co-cultured with 9.9 × 107 CFU of E. coli DH5α at 28 days of life; vaccinated + infected birds (n = 9) were inoculated with 6 × 105 cells of attenuated H. meleagridis (passage 302) co-cultured with 1.2 × 108 CFU of E. coli DH5α and 6 × 105 cells of virulent H. meleagridis (passage 28) co-cultured with 9.9 × 107 CFU of E. coli DH5α at 1 and 28 days of life, respectively. All inocula were administered via oral and cloacal routes, and the procedures were followed as described previously [29]. The birds were examined daily for clinical signs. Necropsy and sampling were performed in three birds from each group at 7 and 14 days post H. meleagridis infection (dpi), with an exception at 7 dpi when only two birds were killed from the vaccinated + infected birds. Finally, at 21 dpi, the remaining three birds from each group were killed and sampled, except from the infected group where no birds were left for necropsy.
Macroscopic lesions in caeca
During necropsy, macroscopic lesions in caeca were recorded and lesion scores (LSs) were assigned based on the previously published scoring scheme ranging from 0 to 4 [30].
Microscopic examination
Caecal tissues from all birds were fixed in 10% neutral buffered formalin, dehydrated, embedded in paraffin, sectioned at 5 µm thickness and stained with hematoxylin and eosin.
Bacteriology
For the quantification of bacterial load of E. coli in caecum, liver and spleen, samples were collected from all birds during necropsy, homogenized and tenfold serial dilution suspensions in phosphate-buffered saline (PBS) were plated on MacConkey agar (Neogen, Heywood, UK) plates in duplicates. Following incubation of plates at 37 °C for 24 h, E. coli CFUs were counted and the bacterial loads were calculated as CFU/g of respective organs.
Microbiota analysis
Caecal contents from each bird were collected in sterile 1.5 mL tubes during necropsy. In case lesions were confined to one side, caecal content of the affected caeca was collected. All samples were stored at −80 °C until further processing. Caecal content was homogenized in a MagNALyzer (Roche). Following homogenization, the DNA was extracted using a QIAamp DNA Stool Mini Kit according to the manufacturer’s instructions (Qiagen, Germany). The DNA concentration was determined with a spectrophotometer and samples diluted to 5 ng/mL were used as a template in PCR with forward primer 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-MID-GT-CCTACGGGNGGCWGCAG-3′ and reverse primer 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-MID-GT-GACTACHVGGGTATCTAATCC-3′. The sequences in italics served for index ligation whereas the underlined sequences allowed for amplification over the V3/V4 region of 16S rRNA genes. MIDs represent different sequences of 5, 6, 7, or 9 base pairs in length which were used to identify individual samples after the whole sequencing run. PCR amplification was performed using a KAPA HiFi Hot Start Ready Mix kit (Kapa Biosystems) and the resulting PCR products were purified using AMPure beads. In the next step, the PCR product concentration was determined with a spectrophotometer before the DNA was diluted to 100 ng/μL and groups of 14 PCR products with different MID sequences were indexed with the same index from Nextera XT Index Kit following the manufacturer’s instructions (Illumina). The next set of 14 PCR products with different MID sequences were indexed with the next index from the Nextera XT Index kit thus allowing an increase in the number of samples analyzed in a single sequencing run. Prior to sequencing, the concentration of differently indexed samples was determined using a KAPA Library Quantification Complete kit (Kapa Biosystems). All indexed samples were diluted to 4 ng/μL and 20 pM phiX DNA was added to a final concentration of 5% (v/v). Sequencing was performed using MiSeq Reagent Kit v3 (600 cycle) and MiSeq apparatus according to the manufacturer’s instructions (Illumina). Quality trimming of the raw reads was performed using TrimmomaticPE v0.32 with sliding window 4 bp and quality read score equal or higher than 20 [31]. Minimal read length must have been at least 150 bp. The fastq files generated after quality trimming were uploaded into QIIME software [32]. Forward and reverse sequences were joined and in the next step chimeric sequences were predicted and excluded by the slayer algorithm. The resulting sequences were then classified by RDP Seqmatch with an OTU (operational taxonomic unit) discrimination level set to 97%.
Immunohistochemistry
In order to investigate the colonization and penetration of caecal tissues by E. coli, paraffin embedded samples of caeca from all birds were processed for immunohistochemistry. Further, to validate the findings from the actual study, caeca of 14 turkeys from infected groups with monoxenic (n = 8) or xenic (n = 6) cultures of H. meleagridis and control birds (n = 4) from previously published experiments were included [29, 33]. Likewise, caecal samples of naturally infected turkeys showing severe fibrinous typhlitis from two field cases were also considered. The IHC protocol for the detection of E. coli was followed as described previously [4]. Briefly, tissue sections mounted on charged glass slides were dewaxed, rehydrated and incubated overnight with a primary monoclonal antibody (anti-E. coli LPS antibody (2D7/1), ab35654, Abcam, Austria). Following incubation, slides were washed with PBS and biotinylated anti-mouse IgG antibody (Vector Laboratories, Austria) was added. Then the vectastain ABC Kit and DAB substrate kit (Vector Laboratories) were used for visualizing the bound antibody. Finally, the sections were counter-stained with Mayer's haematoxylin (Merck KGaA, Austria) and observed under a microscope.
Statistical analysis
Data of alpha diversity matrices and E. coli count were analyzed by one-way ANOVA followed by Tukey’s multiple comparison post hoc test. The Spearman’s rank correlation coefficient (r) was used to evaluate relationship between caecal LSs and E. coli count in the caecum. All analyses were performed in SPSS (IBM® SPSS® version 25; IBM cooperation, New York, USA). The p value < 0.05 was considered as statistically significant.
Results
Clinical signs
One bird from the vaccinated + infected group showed swollen hock joints and was euthanized before scheduled killing. No clinical signs were seen in rest of birds.
Macroscopic lesions
The early-euthanized bird with swollen hock joints showed arthritis. The control birds that were not inoculated with H. meleagridis were devoid of pathological lesions similar to those which received attenuated histomonads (LS 0, Figure 1A). Following inoculation of birds with attenuated and/or virulent H. meleagridis strains, severity of caecal lesions varied, and LS 0, 1, 3 and 4 were recorded (Figures 1B–D). Therefore, birds were categorized into four groups according to their inoculation status and severity of caecal lesions for the analysis of sequencing, bacteriology and IHC data (Table 2). These groups were as follows: (i) negative control (n = 9; birds not inoculated with H. meleagridis, LS 0), (ii) no lesion (n = 11; birds inoculated with H. meleagridis that showed LS 0), (iii) mild lesion (n = 6; birds inoculated with H. meleagridis that showed LS 1), and (iv) severe lesion (n = 6; birds inoculated with H. meleagridis that showed LS 3 or 4). Negative control birds were separated from H. meleagridis inoculated birds with no lesion to observe possible differences induced by parasite inoculation. The presence of compact fibrinous mass in the lumen of one or both caeca are allocated in the macroscopic LS 3 or LS 4 [30]. As the samples were always taken from the affected caecum, birds showing LS 3 and 4 were grouped together.
Macroscopic lesions in caecum of turkeys in different groups. A Negative control group, not inoculated with H. meleagridis; B no lesion group, inoculated with H. meleagridis; C mild lesion group, inoculated with H. meleagridis (hemorrhages are shown with arrow heads); D severe lesion group, inoculated with H. meleagridis.
Histopathology
No histological lesions were found in caeca of birds from negative control and no lesion groups (Figures 2A and B). Birds belonging to the mild lesion group showed mucosal erosion with heterophilic/eosinophilic infiltration limited to lamina propria and submucosa (Figure 2C). Severe necrosis in mucosa with inflammatory cells extended transmurally was observed in birds assigned to the severe lesion group (Figure 2D).
Histopathology in caecum of turkeys in different groups. A Negative control group, not inoculated with H. meleagridis, tissues are normal; B no lesion group, inoculated with H. meleagridis, tissues are normal; C mild lesion group, inoculated with H. meleagridis, erosion of mucosal epithelium with heterophilic/eosinophilic cells infiltration extended to submucosa; D severe lesion group, inoculated with H. meleagridis, extensive necrosis with pyogranulomatous reaction (asterisk) and transmural infiltration of inflammatory cells.
Bacteriology
Pure colonies of Streptococcus sp. were isolated from the affected joint of the early euthanized bird. Average bacterial counts of E. coli in caeca of birds from different groups ranged from 8 to 9 log CFU/g and differences among groups were not statistically significant (Figure 3A). Although a very slight tendency of positive correlation was observed between the severity of caecal macroscopic LSs and the E. coli load but this was not statistically significant (r = 0.11; Figure 3B). In systemic organs, one bird from the negative control group was positive in spleen and contained 1.56 log CFU/g of E. coli. In another bird from the no lesion group that was inoculated with attenuated H. meleagridis, liver and spleen were found positive with 3.1 log CFU/g and 3 log CFU/g of E. coli, respectively.
Average E. coli counts (A) and their correlation with the severity of macroscopic lesions (B) in caeca of turkeys. A Groups are as follows: negative control (birds not inoculated with H. meleagridis), no lesion (birds inoculated with H. meleagridis that showed no lesions), mild lesion (birds inoculated with H. meleagridis that showed LS 1), severe lesion (birds inoculated with H. meleagridis that showed LS 3 or 4). Results are expressed as mean±SEM, values with different letters are statistically significant; B Spearman’s correlation coefficient of caecal lesion score with bacterial load of E. coli count in caeca. Lesion scores were assigned based on the previously published scoring scheme [31].
Caecal microbiota
The OTUs identified in caecal samples were classified into 13 bacterial phyla and one archaeal phylum. Overall Firmicutes (78.2%), Bacteroidetes (13.3%), Acidobacteria (7.4%), Verrucomicrobia (4.8%) and Proteobacteria (3.4%) were the most dominant phyla (Figure 4).
Relative abundance (%) of OTUs representing the caecal microbiota at phylum level in turkeys. Groups are as follows: negative control (birds not inoculated with H. meleagridis), no lesion (birds inoculated with H. meleagridis that showed no lesions), mild lesion (birds inoculated H. meleagridis that showed LS 1), severe lesion (birds inoculated with H. meleagridis that showed LS 3 or 4).
Bacterial diversity and richness was evaluated among the four groups (negative control, no lesion, mild lesion and severe lesion) using alpha diversity indices (Observed species, Chao 1, Shannon index and Equability). Bacterial diversity and richness significantly decreased in birds of the severe lesion group compared to others (Figures 5A–D).
Alpha diversity indices of microbial community in caeca of turkeys. A Observed species, estimates the total number of OTUs; B Chao1, estimates the OTUs richness; C shannon index, an estimation of microbial diversity; D equability, estimates OTUs evenness. Groups are as follows: negative control (birds not inoculated with H. meleagridis), no lesion (birds inoculated with H. meleagridis that showed no lesions), mild lesion (birds inoculated with H. meleagridis that showed LS 1), severe lesion (birds inoculated with H. meleagridis that showed LS 3 or 4). Results are shown as mean ± SEM. Different letters denote statistically significant differences.
The relative abundance of OTUs was analysed at species level and, where possible, confirmation of species was done by aligning with available sequences using nucleotide BLAST or RNA/ITS databases. Out of 15 most abundant bacterial species, the relative abundance of Anaerotignum lactatifermentans, E. coli, Faecalibacterium prausnitzii, Akkermansia muciniphila, Caecibacterium sporoformans, Clostridium leptum and Butyricicoccus pullicaecorum were more abundant in birds of the severe lesion group in comparison to other groups. In contrary, Alistipes putredinis, Streptococcus alactolyticus, Lactobacillus salivarius, Lactobacillus reuteri were less abundant in birds with the highest lesion scores (Figure 6).
Mean relative abundance (%) of the 15 most abundant bacterial species in caeca of turkeys. Groups are as follows: negative control (birds not inoculated with H. meleagridis), no lesion (birds inoculated with H. meleagridis that showed no lesions), mild lesion (birds inoculated with H. meleagridis that showed LS 1), severe lesion (birds inoculated with H. meleagridis that showed LS 3 or 4).
Immunohistochemistry
E. coli cells were detected in caecal luminal content of one and three birds from the mild lesion and the severe lesion groups, respectively (Table 3). E. coli did not penetrate into the caecal wall of birds from any of the groups (Figures 7A and B). In samples taken from previously published experimental studies [29, 33], E. coli cells were detected in the luminal content of 3 out of 8 turkeys inoculated with monoxenic H. meleagridis culture (Figure 7C) and in 4 out of 6 turkeys inoculated with xenic H. meleagridis culture, in which some E. coli cells were attached on the necrotic epithelium of the caecal mucosa (Figure 7D). None of the samples from both trials showed infiltration of E. coli in the caecal wall. Likewise, caecal content and necrotic epithelium of mucosal surface were positive for E. coli while layers of caecal wall were negative in samples taken from field cases of histomonosis in turkeys (Figure 7E).
Immunohistochemical detection of E. coli in the caecal samples of turkeys. A Negative control, absence of immunostaining for E. coli; B presence of E. coli in the caecal content (arrow heads) of a sample from the severe lesion group that was infected with H. meleagridis; C–E profound presence of E. coli (arrow heads) in the caecal content without any infiltration into the caecal wall, caeca are severely inflamed and necrotized after experimental infection with monoxenic H. meleagridis culture (C), xenic H. meleagridis culture (D) natural infection from a field case E.
Discussion
The gut is the reservoir for E. coli in poultry and the features of the bacterial population in intestine has been largely explored in chickens [2]. It is well known that inflammation and necrosis caused by gut pathogens in chickens can substantially affect the structure of gut microbiota. However, such studies are lacking in turkeys. H. meleagridis is a well-recognized protozoal parasite that can cause severe caecal tissue destruction in turkeys and the relationship between H. meleagridis and E. coli is also evident in vitro [34]. Thus, in the present study, H. meleagridis was taken as a model organism to investigate the influence of pathology-associated changes on the caecal microbiota with particular attention to E. coli. In order to extend this subject, the ability of E. coli to invade the caecal wall was also investigated.
Quantification of the total E. coli counts among different groups showed that there was no association between the load of E. coli and the pathologies in the gut. The finding is in agreement with a previous study in mice where inflammation and disease severity did not influence the number of E. coli [35]. However, a link could be determined between severity of gut inflammation and microbiota composition. The pathology induced by H. meleagridis was associated with the pronounced shift in microbial structure, microbial richness and diversity. Similar effects were noticed in chickens that were either co-infected with H. meleagridis and avian pathogenic E. coli [4] or with E. tenella [36, 37]. In controversy, in a different study with E. tenella infection in broiler chickens, no changes were reported in alpha diversity even in caecal samples with the highest lesion scores [10]. This discrepancy between studies can be due to different factors such as inoculated pathogen, host, age, stocking density or gender; all have been shown to influence the microbiome composition and diversity [38].
The relative abundance of OTUs belonging to Anaerotignum lactatifermentans, E. coli and Faecalibacterium prausnitzii increased in caecal samples of birds allocated in the severe lesion group. The higher relative abundance of Anaerotignum lactatifermentans, amino acid decomposing bacterium, in birds with severe lesions may be attributed to the fibrinous necrotic enteritis [39, 40]. Fibrinous inflammation usually occurs when vasculature fluid leaks allowing large plasma proteins, especially fibrinogen, to enter the tissue [41]. Furthermore, fibrinonecrotic compact mass in caecum blocks the digesta to pass from small intestine and this might result into increased retention time and ultimately flourish the population of proteolytic bacteria [42].
The increase in relative abundance of phylum Proteobacteria, in particular E. coli population, in birds of the severe lesion group is obviously an indication of gut dysbiosis induced by inflammation and necrosis. However, the high throughput sequencing identifies the overall microbial communities and their relative abundance to each other, in which every decrease in one taxon’s abundance leads to an equivalent increase across the remaining taxa while CFU count determines the absolute bacterial load. Therefore, building up biological interpretations depending only on sequencing data might be misleading as the measurement of each taxon’s relative abundance is dependent on the populations of all other taxa. The enrichment in one bacterial taxa (high relative abundance) does not necessarily reflect the outgrowth of this taxa (actual abundance) [43]. The phenomenon is nicely reflected by the absolute and relative population of E. coli in this study. Absolute counts of E. coli did not differ across all groups of birds. Relative increase of E. coli in birds with severe lesions is therefore a consequence of minimizing other taxa and not E. coli overgrowth. Nevertheless, as stated above, it is interesting that E. coli maintains the same population in turkeys with or without gut inflammation. Likewise, the relative abundance of Faecalbacterium prausnitzii, a potentially beneficial microbe was strongly increased in birds belonging to the severe lesion group. Increase of Faecalbacterium prausnitzii, was associated also with an increase of closely related Fournierella massiliensis suggesting that the observation was not an artefact. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacteria [44] thus, the increased relative abundance of this species in the condition with the highest lesion score might be due to a beneficial response against inflammation [45]. A specific feature of Faecalibacterium prausnitzii is the loss of spore formation [46, 47] with alternative measures to cope with oxygen. Faecalibacterium prausnitzii is capable to grow at microaerophilic conditions [48] which may enable this bacterium to survive local inflammation associated with generation of oxygen radicals by macrophages and heterophils. Moreover, its ability to produce butyrate and its location in the mucous layer near the epithelial cells [49] might enable it to share an ecological niche with E. coli and also supress expansion of pathosymbionts to colonize the mucus layer, however it needs to be further investigated in details.
Lactobacillus as a probiotic has been shown to control disease aggravation via modulation of the innate and acquired immune system [50, 51]. Therefore, the high abundance of Lactobacillus salivarius and reuteri in relatively healthy birds (negative control, no lesion and mild lesion groups) compared to those in severe lesion group with LS 3 or 4 might have contributed to an early immune response, reducing the invasion of H. meleagridis and opportunistic pathogenic bacteria along epithelial cells. Lactobacilli were observed to be the predominant microbes in the small intestine of chickens and turkeys [52, 53]. Therefore, decreased Lactobacillus abundance in birds with typhlitis could also be due to the blockage of digesta from small intestine because of compact fibrino necrotic mass in the caecum. The same explanation can be stated for Streptococcus alactolyticus, which was also detected with high relative abundance in negative control birds and abundance decreased in infected birds with lesions.
With IHC, E. coli could only be detected in luminal content of caecum and preserved caecal content (coagula). Low numbers of E. coli positive caecal samples in the actual study can be explained by the fact that routine steps in tissue processing for paraffin sections might remove contents from the gut as previously reported [54]. Anyhow, it is interesting to observe that E. coli cells did not infiltrate into the caecal wall even in birds with high relative abundance of E. coli and severe sloughing of mucosal epithelium. Similar findings in turkeys from other experimental studies and field cases showed that the pattern remains the same in birds experimentally infected with monoxenic or xenic H. meleagridis cultures or in those that are naturally infected. Altogether, it was demonstrated that E. coli in the gut of turkeys do not show a tendency to penetrate the gut wall and the effect was not potentiated by the presence of inflammation and necrosis. Very limited re-isolation of E. coli from systemic organs with no definite pattern based on severity of lesions indicated absence of systemic translocation of E. coli from the gut.
The significant dysbiosis in the caecal microbiota of turkeys was correlated with the severity of pathology following H. meleagridis infection. This perturbation was associated with increased relative abundance but not an absolute count of E. coli in the gut. Inflammation and necrosis due to H. meleagridis did not provoke penetration of caecal wall and systemic translocation of E. coli from the gut. Thus, intestinal E. coli cells in turkeys do not seem to have a tendency to cause systemic bacteremia even when the gut barrier is compromised.
Availability of data and materials
Raw sequence reads were uploaded to the NCBI BioProject databank (PRJNA718176).
Change history
03 November 2021
Funding section has been updated.
Abbreviations
- CFU:
-
Colony forming units
- Dpi:
-
Days post H. meleagridis infection
- E. coli :
-
Escherichia coli
- H. meleagridis :
-
Histomonas meleagridis
- E. tenella :
-
Eimeria tenella
- IHC:
-
Immunohistochemistry
- LS:
-
Lesion score
- OTU:
-
Operational taxonomic unit
- PBS:
-
Phosphate buffered saline
- PCR:
-
Polymerase chain reaction
References
Diether NE, Willing BP (2019) Microbial fermentation of dietary protein: an important factor in diet–microbe–host interaction. Microorganisms 7:19. https://doi.org/10.3390/microorganisms7010019
Rychlik I (2020) Composition and function of chicken gut microbiota. Animals 10:103. https://doi.org/10.3390/ani10010103
Diaz Carrasco JM, Casanova NA, Fernández Miyakawa ME (2019) Microbiota, gut health and chicken productivity: what is the connection? Microorganisms 7:374. https://doi.org/10.3390/microorganisms7100374
Abdelhamid MK, Quijada NM, Dzieciol M, Hatfaludi T, Bilic I, Selberherr E, Liebhart D, Hess C, Hess M, Paudel S (2020) Co-infection of chicken layers with Histomonas meleagridis and avian pathogenic Escherichia coli is associated with dysbiosis, cecal colonization and translocation of the bacteria from the gut lumen. Front Microbiol 11:586437. https://doi.org/10.3389/fmicb.2020.586437
Ruhnau D, Hess C, Grenier B, Doupovec B, Schatzmayr D, Hess M, Awad WA (2020) The Mycotoxin Deoxynivalenol (DON) promotes Campylobacter jejuni multiplication in the intestine of broiler chickens with consequences on bacterial translocation and gut integrity. Front Vet Sci 7:1027. https://doi.org/10.3389/fvets.2020.573894
Shang Y, Kumar S, Oakley B, Kim WK (2018) Chicken gut microbiota: importance and detection technology. Front Vet Sci 5:254. https://doi.org/10.3389/fvets.2018.00254
Kempf F, Menanteau P, Rychlik I, Kubasová T, Trotereau J, Virlogeux-Payant I, Schaeffer S, Schouler C, Drumo R, Guitton E (2020) Gut microbiota composition before infection determines the Salmonella super-and low-shedder phenotypes in chicken. Microb Biotechnol 13:1611–1630. https://doi.org/10.1111/1751-7915.13621
Hankel J, Jung K, Kuder H, Keller B, Keller C, Galvez E, Strowig T, Visscher C (2019) Caecal microbiota of experimentally Campylobacter jejuni-infected chickens at different ages. Front Microbiol 10:2303. https://doi.org/10.3389/fmicb.2019.02303
Chen H-L, Zhao X-Y, Zhao G-X, Huang H-B, Li H-R, Shi C-W, Yang W-T, Jiang Y-L, Wang J-Z, Ye L-P (2020) Dissection of the cecal microbial community in chickens after Eimeria tenella infection. Parasit Vectors 13:56. https://doi.org/10.1186/s13071-020-3897-6
Macdonald SE, Nolan MJ, Harman K, Boulton K, Hume DA, Tomley FM, Stabler RA, Blake DP (2017) Effects of Eimeria tenella infection on chicken caecal microbiome diversity, exploring variation associated with severity of pathology. PLoS One 12:e0184890. https://doi.org/10.1371/journal.pone.0184890
Wilkinson TJ, Cowan A, Vallin H, Onime L, Oyama LB, Cameron S, Gonot C, Moorby J, Waddams K, Theobald V (2017) Characterization of the microbiome along the gastrointestinal tract of growing turkeys. Front Microbiol 8:1089. https://doi.org/10.3389/fmicb.2017.01089
D’Andreano S, Bonastre AS, Francino O, Martí AC, Lecchi C, Grilli G, Giovanardi D, Ceciliani F (2017) Gastrointestinal microbial population of turkey (Meleagris gallopavo) affected by hemorrhagic enteritis virus. Poult Sci 96:3550–3558. https://doi.org/10.3382/ps/pex139
Danzeisen JL, Clayton JB, Huang H, Knights D, McComb B, Hayer SS, Johnson TJ (2015) Temporal relationships exist between cecum, ileum, and litter bacterial microbiomes in a commercial turkey flock, and subtherapeutic penicillin treatment impacts ileum bacterial community establishment. Front Vet Sci 2:56. https://doi.org/10.3389/fvets.2015.00056
Adhikari B, Tellez-Isaias G, Jiang T, Wooming B, Kwon YM (2020) Comprehensive survey of the litter bacterial communities in commercial turkey farms. Front Vet Sci 7:596933. https://doi.org/10.3389/fvets.2020.596933
Taylor KJ, Ngunjiri JM, Abundo MC, Jang H, Elaish M, Ghorbani A, Mahesh K, Weber BP, Johnson TJ, Lee C-W (2020) Respiratory and gut microbiota in commercial turkey flocks with disparate weight gain trajectories display differential compositional dynamics. Appl Environ Microbiol 86:e00431-20. https://doi.org/10.1128/AEM.00431-20
Gonzalez-Ortiz G, Olukosi O, Jurgens G, Apajalahti J, Bedford M (2020) Short-chain fatty acids and ceca microbiota profiles in broilers and turkeys in response to diets supplemented with phytase at varying concentrations, with or without xylanase. Poult Sci 99:2068–2077. https://doi.org/10.1016/j.psj.2019.11.051
Danzeisen JL, Calvert AJ, Noll SL, McComb B, Sherwood JS, Logue CM, Johnson TJ (2013) Succession of the turkey gastrointestinal bacterial microbiome related to weight gain. PeerJ 1:e237. https://doi.org/10.7717/peerj.237
Ballou AL, Ali RA, Mendoza MA, Ellis J, Hassan HM, Croom W, Koci MD (2016) Development of the chick microbiome: how early exposure influences future microbial diversity. Front Vet Sci 3:2. https://doi.org/10.3389/fvets.2016.00002
Leitner G, Heller E (1992) Colonization of Escherichia coli in young turkeys and chickens. Avian Dis 36:211–220
Hafez HM (2005) Current knowledge and prospective risk analysis related to ongoing turkey diseaseIn. In: Proceedings of 14 international congress of the World Poultry Association. Istanbul, Turkey, pp 138-149
Sárközy G, Semjén G, Laczay P, Horváth E, Schmidt J (2002) Pulse and continuous oral norfloxacin treatment of experimentally induced Escherichia coli infection in broiler chicks and turkey poults. Acta Vet Acad Sci Hung 50:199–210. https://doi.org/10.1556/AVet.50.2002.2.9
De Oliveira AL, Newman DM, Sato Y, Noel A, Rauk B, Nolan LK, Barbieri NL, Logue CM (2020) Characterization of avian pathogenic Escherichia coli (APEC) associated with turkey cellulitis in Iowa. Front Vet Sci 7:380. https://doi.org/10.3389/fvets.2020.00380
Collingwood C, Kemmett K, Williams N, Wigley P (2014) Is the concept of avian pathogenic Escherichia coli as a single pathotype fundamentally flawed? Front Vet Sci 1:5. https://doi.org/10.3389/fvets.2014.00005
Huff G, Huff W, Balog J, Rath N, Anthony N, Nestor K (2005) Stress response differences and disease susceptibility reflected by heterophil to lymphocyte ratio in turkeys selected for increased body weight. Poult Sci 84:709–717. https://doi.org/10.1093/ps/84.5.709
Huff G, Huff W, Rath N, Balog J, Anthony N, Nestor K (2006) Stress-induced colibacillosis and turkey osteomyelitis complex in turkeys selected for increased body weight. Poult Sci 85:266–272. https://doi.org/10.1093/ps/85.2.266
Umar S, Delverdier M, Delpont M, Belkasmi SF, Teillaud A, Bleuart C, Pardo I, El Houadfi M, Guérin J-L, Ducatez MF (2018) Co-infection of turkeys with Escherichia coli (O78) and H6N1 avian influenza virus. Avian Pathol 47:314–324. https://doi.org/10.1080/03079457.2018.1449942
Turpin EA, Perkins LE, Swayne DE (2002) Experimental infection of turkeys with avian pneumovirus and either newcastle disease virus or Escherichia coli. Avian Dis 46:412–422. https://doi.org/10.1637/0005-2086(2002)046[0412:EIOTWA]2.0.CO;2
Ganas P, Liebhart D, Glösmann M, Hess C, Hess M (2012) Escherichia coli strongly supports the growth of Histomonas meleagridis, in a monoxenic culture, without influence on its pathogenicity. Int J Parasitol 42:893–901. https://doi.org/10.1016/j.ijpara.2012.07.007
Lagler J, Schmidt S, Mitra T, Stadler M, Wernsdorf P, Grafl B, Hatfaludi T, Hess M, Gerner W, Liebhart D (2021) Comparative investigation of IFN-γ-producing T cells in chickens and turkeys following vaccination and infection with the extracellular parasite Histomonas meleagridis. Dev Comp Immunol 116:103949. https://doi.org/10.1016/j.dci.2020.103949
Windisch M, Hess M (2010) Experimental infection of chickens with Histomonas meleagridis confirms the presence of antibodies in different parts of the intestine. Parasite Immunol 32:29–35. https://doi.org/10.1111/j.1365-3024.2009.01159.x
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Kidane FA, Mitra T, Wernsdorf P, Hess M, Liebhart D (2018) Allocation of interferon gamma mrna positive cells in caecum hallmarks a protective trait against histomonosis. Front Immunol 9:1164. https://doi.org/10.3389/fimmu.2018.01164
Bilic I, Hess M (2020) Interplay between Histomonas meleagridis and bacteria: mutualistic or predator–prey? Trends Parasitol 36:232–235. https://doi.org/10.1016/j.pt.2019.12.015
Kittana H, Gomes-Neto JC, Heck K, Geis AL, Segura Muñoz RR, Cody LA, Schmaltz RJ, Bindels LB, Sinha R, Hostetter JM (2018) Commensal Escherichia coli strains can promote intestinal inflammation via differential interleukin-6 production. Front Immunol 9:2318. https://doi.org/10.3389/fimmu.2018.02318
Zhou B-h, Jia L-s, Wei S-s, Ding H-y, Yang J-y, Wang H-w (2020) Effects of Eimeria tenella infection on the barrier damage and microbiota diversity of chicken cecum. Poult Sci 99:1297–1305. https://doi.org/10.1016/j.psj.2019.10.073
Huang G, Tang X, Bi F, Hao Z, Han Z, Suo J, Zhang S, Wang S, Duan C, Yu Z (2018) Eimeria tenella infection perturbs the chicken gut microbiota from the onset of oocyst shedding. Vet Parasitol 258:30–37. https://doi.org/10.1016/j.vetpar.2018.06.005
Kers JG, Velkers FC, Fischer EA, Hermes GD, Stegeman JA, Smidt H (2018) Host and environmental factors affecting the intestinal microbiota in chickens. Front Microbiol 9:235. https://doi.org/10.3389/fmicb.2018.00235
Ueki A, Goto K, Ohtaki Y, Kaku N, Ueki K (2017) Description of Anaerotignum aminivorans gen. nov., sp. nov., a strictly anaerobic, amino-acid-decomposing bacterium isolated from a methanogenic reactor, and reclassification of Clostridium propionicum, Clostridium neopropionicum and Clostridium lactatifermentans as species of the genus Anaerotignum. Int J Syst Evol Microbiol 67:4146–4153. https://doi.org/10.1099/ijsem.0.002268
Sentíes-Cué G, Chin R, Shivaprasad H (2009) Systemic histomoniasis associated with high mortality and unusual lesions in the bursa of Fabricius, kidneys, and lungs in commercial turkeys. Avian Dis 53:231–238. https://doi.org/10.1637/8363-051508-Reg.1
Hoda SA, Cheng E (2017) Robbins basic pathology. Am J Clin Pathol 148:557. https://doi.org/10.1093/ajcp/aqx095
Macfarlane G, Cummings J, Macfarlane S, Gibson G (1989) Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous culture system. J Appl Microbiol 67:521–527
Hartley MG, Ralph E, Norville IH, Prior JL, Atkins TP (2019) Comparison of PCR and viable count as a method for enumeration of bacteria in an A/J mouse aerosol model of Q Fever. Front Microbiol 10:1552. https://doi.org/10.3389/fmicb.2019.01552
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Natl Acad Sci 105:16731–16736
Yan W, Sun C, Yuan J, Yang N (2017) Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci Rep 7:45308. https://doi.org/10.1038/srep45308
Galperin MY (2013) Genome diversity of spore-forming firmicutes. Microbiol Spectr. https://doi.org/10.1128/microbiolspectrum.TBS-0015-2012
Medvecky M, Cejkova D, Polansky O, Karasova D, Kubasova T, Cizek A, Rychlik I (2018) Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genom 19:561. https://doi.org/10.1186/s12864-018-4959-4
Khan MT, Duncan SH, Stams AJ, Van Dijl JM, Flint HJ, Harmsen HJ (2012) The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J 6:1578–1585. https://doi.org/10.1038/ismej.2012.5
Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ (2002) Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52:2141–2146. https://doi.org/10.1099/00207713-52-6-2141
Hessenberger S, Schatzmayr G, Teichmann K (2016) In vitro inhibition of Eimeria tenella sporozoite invasion into host cells by probiotics. Vet Parasitol 229:93–98. https://doi.org/10.1016/j.vetpar.2016.10.001
Dalloul RA, Lillehoj HS, Tamim NM, Shellem TA, Doerr JA (2005) Induction of local protective immunity to Eimeria acervulina by a Lactobacillus-based probiotic. Comp Immunol Microbiol Infect Dis 28:351–361. https://doi.org/10.1016/j.cimid.2005.09.001
Gong J, Si W, Forster RJ, Huang R, Yu H, Yin Y, Yang C, Han Y (2007) 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol Ecol 59:147–157. https://doi.org/10.1111/j.1574-6941.2006.00193.x
Johnson TA, Sylte MJ, Looft T (2019) In-feed bacitracin methylene disalicylate modulates the turkey microbiota and metabolome in a dose-dependent manner. Sci Rep 9:8212. https://doi.org/10.1038/s41598-019-44338-5
Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR (2004) Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol 28:23–27
Acknowledgments
The authors are thankful to Delfina Jandreski-Cvetkovic and Claudia Ibesich for their technical input. We also thank Fana Alem Kidane for providing some samples for immunohistochemistry.
Funding
The study was partially funded by the European Regional Development Fund, Cross-border Cooperation Programme INTERREG V-A Austria-Czech Republic of the European Union, project number: ATCZ194, INPOMED (Innovations in Poultry Medicine). Open access funding provided by University of Veterinary Medicine Vienna.
Author information
Authors and Affiliations
Contributions
CH, DL, MH and SP conceived and designed the study; MKA, CH, JL, DL and SP performed the animal trial and sampling; TH prepared the H. meleagridis cultures; MKA, CH and SP performed the bacteriological investigation; MKA, DL and SP performed the histological investigation; IR, MC and DK prepared the samples and conducted sequencing analysis; MKA and SP prepared the first draft and all the authors provided their inputs in the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The trial was approved by the institutional ethics and animal welfare committee and the national authority according to §§ 26ff. of Animal Experiments Act, Tierversuchsgesetz 2012–TVG 2012 (License Number GZ.68.205/0161-WF/V/3b/2017).
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Abdelhamid, M.K., Rychlik, I., Hess, C. et al. Typhlitis induced by Histomonas meleagridis affects relative but not the absolute Escherichia coli counts and invasion in the gut in turkeys. Vet Res 52, 92 (2021). https://doi.org/10.1186/s13567-021-00962-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-021-00962-6