Abstract
Background
With COVID-19 still in its pandemic stage, extensive research has generated increasing amounts of data and knowledge. As many studies are published within a short span of time, we often lose an integrative and comprehensive picture of host-coronavirus interaction (HCI) mechanisms. As of early April 2021, the ImmPort database has stored 7 studies (with 6 having details) that cover topics including molecular immune signatures, epitopes, and sex differences in terms of mortality in COVID-19 patients. The Coronavirus Infectious Disease Ontology (CIDO) represents basic HCI information. We hypothesize that the CIDO can be used as the platform to represent newly recorded information from ImmPort leading the reinforcement of CIDO.
Methods
The CIDO was used as the semantic platform for logically modeling and representing newly identified knowledge reported in the 6 ImmPort studies. A recursive eXtensible Ontology Development (XOD) strategy was established to support the CIDO representation and enhancement. Secondary data analysis was also performed to analyze different aspects of the HCI from these ImmPort studies and other related literature reports.
Results
The topics covered by the 6 ImmPort papers were identified to overlap with existing CIDO representation. SARS-CoV-2 viral S protein related HCI knowledge was emphasized for CIDO modeling, including its binding with ACE2, mutations causing different variants, and epitope homology by comparison with other coronavirus S proteins. Different types of cytokine signatures were also identified and added to CIDO. Our secondary analysis of two cohort COVID-19 studies with cytokine panel detection found that a total of 11 cytokines were up-regulated in female patients after infection and 8 cytokines in male patients. These sex-specific gene responses were newly modeled and represented in CIDO. A new DL query was generated to demonstrate the benefits of such integrative ontology representation. Furthermore, IL-10 signaling pathway was found to be statistically significant for both male patients and female patients.
Conclusion
Using the recursive XOD strategy, six new ImmPort COVID-19 studies were systematically reviewed, the results were modeled and represented in CIDO, leading to the enhancement of CIDO. The enhanced ontology and further seconary analysis supported more comprehensive understanding of the molecular mechanism of host responses to COVID-19 infection.
Similar content being viewed by others
Background
COVID-19 has posed a series of major crises in global public health. With more than 20,000 confirmed cases and almost 1000 deaths in the European Region on March 12, 2020, the WHO declared the pandemic status of the COVID-19 outbreak. As of May 3, 2021, the COVID-19 pandemic had caused over 140 million confirmed cases with over 3 million deaths worldwide, and 31 million confirmed cases with 565 K deaths in the USA [1]. It is critical to systematically study the molecular mechanisms of COVID-19 disease formation and host responses in order to fully understand, prevent, and treat COVID-19.
To better study and understand the disease mechanism, extensive research has been conducted in a relatively short period of time. With tens of thousands of papers published on host-coronavirus interactions (HCIs), a major bottleneck is how to incorporate all the studies into a more comprehensive understanding of the HCI mechanisms. For example, the ImmPort database provides the data related to immune responses stimulated by various agents including infections and vaccines [2]. As of April 22, 2021, ImmPort has included 7 studies on COVID-19, and 6 of these studies have included unique and large data sets.
The Coronavirus Infectious Disease Ontology (CIDO) is a community-based ontology in the domain of coronaviruses [3]. CIDO covers various coronavirus infectious diseases, with a major focus on COVID-19. The areas of CIDO coverage are broad, including various coronaviruses, hosts, host-coronavirus interactions, phenotypes, vaccine, drugs, epidemiology, etc. As a formal biomedical ontology, CIDO is a human- and computer-interpretable representation of the entities and relationships among the entities in the specific coronavirus infectious disease domain. As of the end of March 2021, the CIDO version 1.0.187 includes over 8111 terms and is continuously updating. Like other ontologies, CIDO allows semantical reasoning and enables humans and machines to make mutually understandable logical inferences. With more studies conducted, it is required to continuously update CIDO. Although manual ontology updates can be time-consuming, automatic updates may not be convincing. With regard to COVID-19, vast knowledge has been learned from the literature and high throughput data analysis. Now the challenge becomes how to keep updating CIDO to have it remain up to date.
There have been different strategies proposed in terms of ontology updating. For example, the Principle of Minimal Change states that the knowledge lost during contraction should be minimal [4]. Solimando and Guerrini propose an ontology adaptation algorithm to fully automatically reformulate ontology axioms to adapt the condition when an entity in the ontology is deleted [5]. A framework of an iterative ontology update with minimal information loss using context-based reasoning method has been proposed [6]. These frameworks and methods emphasize the maintenance of existing ontology information and minimal context loss. To support ontology interoperability, the eXtensible Ontology development (XOD) strategy [7] emphasizes term reuse (instead of regenerating new terms, XOD1) and semantic alignment (XOD2), ontology design pattern usage for new term and axiom addition (XOD3), and community effort (XOD4).
In this paper, we applied the recursive usage of the XOD strategy for our CIDO updates by actively, progressively, and recursively identifying new knowledge from literature mining and manually annotated papers, or from our secondary data analysis of deposited data (ImmPort).
In our study, we extracted or performed secondary analysis on the ImmPort COVID-19 studies, applies the XOD strategy to the model and represent the experimentally verified results in the CIDO as a way to update and enhance CIDO. We also performed modeling and represent the learned knowledge in ontology. The whole process has been recursive because we do it periodically and consistently. Every time we do, we will improve the ontology through our recursive XOD-based ontology development and modeling.
Our study focused on the spike glycoprotein (S protein) and comparison in male and female responses to COVID-19. We include these two use cases because of their importance for coronavirus-host interactions and in disease outcome, respectively. The S protein of the SARS-CoV-2 plays a critical role in host-coronavirus interactions. The initial entry of the virus is driven predominantly by the S protein [8] which binds to the host cell’s ACE2 to initiate viral infection. This process is aided by transmembrane protease serine 2 (i.e., TMPRSS2) [9]. For this reason, S protein has been chosen as an antigen for several approved vaccines worldwide [10]. Additionally, males have consistently shown to have higher mortality and hospitalization rates in comparison to females [11]. We are using male and female as synonymous for biological sex unless otherwise specified as gender.
Methods
ImmPort secondary data analysis
The COVID-19 related papers were obtained from the ImmPort website (https://www.immport.org/shared/home). For each ImmPort COVID-19 study, ImmPort provides a list of structured information including a summary, design, adverse event, assessment, etc. Links to PubMed are also provided. All collected information was annotated, and a secondary data analysis was performed.
Gene pathway analysis of sex comparison using ImmPort data using Reactome
The differentially expressed proteins in female or male COVID-19 patients compared to healthy controls were collected from Takahashi et al. [12] and mapped to a corresponding gene ID from a background list of their cytokine assay using binomial distribution. These genes cover different interferons, cytokines, chemokines, and growth factors. A significance cutoff of p < =0.05 was applied to the study results by secondary data analysis as shown in Extended Table 1 or Extended Table 2. The NCBI Gene names were then used in the Reactome pathway browser to identify the enriched pathways based on female or male gene lists.
Recursive CIDO ontology updating of host-coronavirus interactions
Our CIDO ontology updating of host-coronavirus interactions followed a recursive eXtensible Ontology development (XOD) strategy [7] as laid out in Fig. 1. Specifically, after new knowledge is learned from the literature or secondary analysis, terms and axioms related to the knowledge are first identified. If the terms are not yet in the CIDO, we will: (i) identify, extract, and reuse the terms from existing ontologies (XOD1), and align them under CIDO (XOD2), (ii) generate new ontology terms using ontology design patterns (XOD3), and or (iii) manually add the terms to CIDO and align them to the semantic structure of existing CIDO (XOD2). Community-based discussion is also involved during the ontology development (XOD4). The XOD processes are done in a recursive way since new knowledge is iteratively and recursively added (Fig. 1).
Strategy of CIDO updating using recursive XOD strategy. The recursive XOD strategy applies the use of the different principles of XOD design for use of new term generation along with creation of new axioms. Further knowledge either from literature or secondary analysis is used as a basis to generate new terms
Based on this strategy, CIDO reuses terms from existing ontologies and aligns all terms within a single semantic framework as defined by the Basic Formal Ontology (BFO) [13]. CIDO follows the Open Biological/Biomedical Ontologies (OBO) Foundry principles [14]. CIDO is an open source project with its source code available at https://github.com/CIDO-ontology/cido. CIDO is released under a Creative Commons 4.0 License. CIDO has been accepted as an OBO library ontology and has been deposited in the Ontobee ontology server [15], BioPortal [16], and OLS [17].
Results
ImmPort data exploration on the basis of existing CIDO development
The method implemented in our study is the recursive XOD strategy (Fig. 1). The added information should be semantically aligned with existing ontology structure. We applied recursive usage of the XOD development pipeline to continuously incorporate and integrate new knowledge data to CIDO.
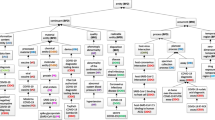
Figure 2 illustrates how CIDO has been developed and how ImmPort data can fit into the existing CIDO structure. A major task addressed in this study was to use the CIDO as the basis, add new knowledge learned from the ImmPort COVID-19 studies (Table 1) to the current version of CIDO, and then perform the secondary analysis to identify new scientific insights about host-SARS-CoV-2 interactions.
CIDO representation of host-coronavirus interactions and the addition of new knowledge learned from papers of ImmPort studies to CIDO. ImmPort studies are labeled with their study IDs (e.g., SDY1667). The termed highlighted are the terms newly added to incorporate the knowledge learned from the corresponding ImmPort studies. This figure shows how CIDO can be updated through our ontology reinforcement strategy
In our study, CIDO is used as a foundation and platform for semantic representation of host responses to COVID-19 infection. CIDO provides basic information about host-coronavirus interactions. Overall, SARS-CoV-2 viral processes include the viral binding to the host cell, entry to the cell, viral genetic replication, assembly of the virion, and release of the virion. CIDO also includes terms and axioms about immune responses to SARS-CoV-2 and has expanded modeling to account for dealing with unique changes from a pandemic. A portion of this information is represented in Fig. 2 showing parts of the SARS-CoV-2 life cycle including viral invasion to the host cell, viral replication, and viral shedding from the cell.
CIDO representation of S protein focused coronavirus invasion, host response, and viral mutation
Modeling of viral invasion and host immune response by S protein
The S protein plays a key role in COVID-19 viral infection and disease pathology. Fig. 3a illustrates how the CIDO ontology represents various coronaviral processes on the surface of and inside the host, such as the main steps of viral infection, reproduction, and shedding (i.e., viral release out of host cell). The process of ‘SARS-CoV-2 S binding to human ACE2’ is defined with the following axioms:
‘has participant’ some ‘ACE2 (human)’
‘has participant’ some ‘S protein of SARS-CoV-2)’
‘part of’ some ‘SARS-CoV-2 entry to human cell’
‘SARS-CoV-2 binding to human cell’
‘SARS-CoV-2 S-ACE2 binding’
Ontological representation of S protein processes and variations. a CIDO ontological representation of viral life cycle in host (cell invasion, genetic replication, and release for cell and invading other cells) of virulent SARS-CoV-2. CIDO ontological representation of coronaviral molecular processes. The right bottom side of the screenshot represents different axioms for the SARS-CoV-2 S binding to human ACE2. b Seven SARS-CoV-2 clades are defined with each has its specific definition. For example, SARS-CoV-2 clade G virus has AA variant S-D614G, which is a variant of S protein of SARS-CoV-2. c CIDO representation of SARS-CoV-2 virus variants such as SARS-CoV-2 Delta variant based on the WHO classification
Such S-ACE2 binding is critical not only to the viral invasion, but also to the manifestation of many COVID-19 phenotypes such as pneumonia, hypertension, heart disease, acute kidney injury [18, 19].
Additionally, the issue of different clades brings about a concern of unique epitopes. A good representative study of their significance for ontology modeling is the study reported in ImmPort (SDY 1667) [20], which found 142 SARS-CoV-2 T-cell epitopes that are homologous epitopes to SARS-CoV-2 and multiple common cold human coronaviruses. Homologous epitopes are defined as any two epitopes, A and B, that exhibit sufficient homology and that when A elicits a host immune response and becomes part of the host immune memory, the A-specific memory will also recognize B. The Immune Epitope Database (IEDB) [21] has collected over 2000 known SARS-CoV-2-specific T or B cell epitopes, and the numbers are being updated every week. A consensus is that IEDB is the proper database to store and maintain the epitopes, and it is inappropriate for CIDO to record all the epitopes. Instead of listing all individual epitopes identified in the ImmPort study (SDY 1667) [20], we propose an ontology design pattern that represents the relation between two proteins that have epitope cross-reactivity. An example of such a representation is shown below:
‘spike glycoprotein (SARS-CoV-2)’: ‘has epitope cross-reactivity’ some ‘spike glycoprotein (HCoV-OC43)’
CIDO modeling of S protein mutation to avoid active host response
SARS-CoV-2 is a virus and by its nature undergoes under selective pressure that results in production of new variations within the viral proteins. For example, the B.1.1.7 clade variant that emerged in the UK [22]. B.1.1.7 already has early evidence for increased transmissibility and potential higher lethality [23]. While there are already ontological representations for immune responses to proteins in specific species, the actual representation of notable protein mutations has not previously been implemented. In Fig. 3a, we provide modeling on how this is done. Each mutant is identified by the protein name followed by a dash and the type of mutation. Identification of these proteins are done at the individual amino acid level as shown below.
‘S-D614G’: ‘variant of’ some ‘spike glycoprotein (SARS-CoV-2)’
S-D614G is interpreted as S protein with a missense mutation that causes the 614th amino acid, aspartic acid (D) to become glycine (G). CIDO has incorporated this to provide a standard set of annotations. A virus variant may have multiple mutations (Fig. 3b), which can also be systematically represented including an axiom as exemplified below:
‘SARS-CoV-2 GRY (B.1.1.7): ‘SARS-CoV-2 clade GR virus’ and (‘has AA variant’ some (S-H69del and S-V70del and S-Y144del and S-N501Y and N-G204R))
One of the challenges for CIDO has been the ontological classification of new SARS-CoV-2 lineages and strains that have emerged. Multiple naming schemas exist, each with different criteria for their categories. The World Health Organization (WHO) has designated certain coronaviruses as either variants of concern or variants of interest and named them using as Alpha, Beta, Gamma, Delta, etc. (Fig. 3c). For example, the SARS-CoV-2 Delta variant is defined as an equivalent axiom:
SARS-CoV-2 Delta variant: ‘SARS-CoV-2 B.1.617.2 virus’ or ('Severe acute respiratory syndrome coronavirus 2’ and (‘derives from’ some ‘SARS-CoV-2 B.1.617.2 virus’))
Here ‘SARS-CoV-2 B.1.617.2 virus’ is a variant classification based on Phylogenetic Assignment of Named Global Outbreak Lineages (PANGO) [24]. The relation ‘derives from’ indicates that the Delta variant includes the SARS-CoV-2 B.1.617.2 virus or any other viral variant derived from the virus. In addition, CIDO also represents the variant classification assigned by the organization of Global Initiative on Sharing Avian Influenza Data (GISAID) [25] (Fig. 3c).
CIDO representation of RAS related drug interruption for treating COVID-19
The ImmPort study (SDY1641) investigated the roles of renin-angiotensin system (RAS) inhibitors, including angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers, in treating COVID-19 patients with hypertension [26]. Patients treated with angiotensin-converting enzyme (ACE) inhibitors or angiotensin II receptor blocker had a lower rate of severe diseases and lowered IL-6 in peripheral blood. The ACE inhibitors or angiotensin II receptor blocker therapy also increased the CD3 and CD8 T cell counts in peripheral blood and decreased the peak of viral load compared to other antihypertensive drugs [26].
RAS is also closely associated with coronavirus S protein since the S protein binds to the host angiotensin-converting-enzyme 2 (ACE2), a key RAS component. The binding between the S glycoprotein and ACE2 needs to be activated by TMPRSS2, a cellular receptor [9] (Fig. 2). Such binding leads to the subsequent downregulation of ACE2 [27, 28]. angiotensin-converting enzyme inhibitors inhibit the activity of ACE, an important component of the RAS that converts angiotensin I to angiotensin II. Therefore, angiotensin-converting enzyme inhibitors decrease the formation of angiotensin II, a vasoconstrictor. Angiotensin II receptor blockers bind to and inhibit the angiotensin II receptor type 1 (AT1), a receptor that has vasoconstriction role. Angiotensin II receptor blockers can then block the activation of the AT1 and prevent the binding of angiotensin II, leading to the treatment of hypertension [29].
We have modeled the above RAS-related process as shown in Fig. 4. Note that many terms and axioms were already represented in our previous CIDO modeling. To add new results obtained from the ImmPort study (SDY1641), multiple new terms and axioms were added as seen in the bold terms in Fig. 4. In our CIDO modeling, we defined many roles, such as ‘ACE Inhibitor role’, ‘angiotensin II receptor blocker role’, ‘vasoconstrictor role’, and ‘vasodilator role’. These roles can be then used to annotate different drugs or molecules, for example:
perindopril: ‘has role’ some ‘ACE inhibitor role’
nifedipine: ‘has role’ some ‘angiotensin II receptor blocker role’
By doing so, the biological relevance of the drugs and molecules can be clearly noted and understood by humans and computers.
CIDO representation of host immune markers between immune profiles and covariates that correlate with COVID-19 outcomes
There are many host immune markers that correlate with COVID-19 outcomes. Figure 4 shows the general pattern of gene expression patterns in an ontological representation of genes (including gene markers) that are susceptible to be up-regulated under a specific condition such as SARS-CoV-2 infection.
Two ImmPort publications from two studies report host immune markers that correlate with COVID-19 outcomes: one that introduce inflammatory cytokine signatures that predicts COVID-19 severity and survival (ImmPort Study SDY1662) [30], and the other that introduces many more immune signatures associated with severe COVID-19 (ImmPort Study SDY1665) [31]. The first paper [30] demonstrates that IL-6 and TNF-alpha both are strong independent predictors of disease severity and death outcomes, with IL-18 also serving as a strong, but not independent predictor. Higher levels of IL-6 elevation are associated with the cytokine release syndrome (CRS), a condition that the SARS-CoV-2 infection also causes in compared to higher immune control [32]. The second paper [31] generated an immune profile by analyzing the immune responses in 113 patients with moderate or severe COVID-19, uncovering an overall increase in innate cell types and a concomitant reduction in T cell number. Severe COVID-19 was found to be associated with the elevation of cytokines and immune pathways associated with type 1 (antiviral), type 2 (anti-helminths), and type 3 (antifungal) type II pathways and higher levels of growth factors, type 1/2/3 cytokines and chemokines. However, patients with moderate COVID-19 had a progressive reduction in type 1 (antiviral) and type 3 (antifungal) responses after an early increase in cytokines and enriched with growth factors [31].
The initial immune signature of IL-6 for COVID-19 disease pathology have been further investigated and associated with other pathologies. For example, COVID-19 is linked to cytokine release syndrome (CRS), and the pathogenesis of CRS is associated with IL-6-mediated production of hyperinflammatory cytokines and plasminogen activator inhibitor-1 (PAI-1) [32]. The inhibition of IL-6 signaling using tocilizumab decreased PAI-1 production and alleviated the clinical symptoms in severe COVID-19 patients [32]. However, Kang et al. [32] also shows that while still elevated compared to healthy control, IL-6, IL-8, and MCP-1 are lower to other CRS diseases. Children had three cytokines increased interferon (IFN)-γ-induced protein 10 (IP10), interleukin (IL)-10 and IL-16 [33].
To model these results, we implemented a new class for biomarker and immune signature. A biomarker is a material entity that has a change in expression associated with a specific response to some specific biological process. An immune signature is a biomarker for some specific disease process. We included new object relations to model these differences for different SARS-CoV-2 disease processes.
IL-6: ‘up-expressed as immune signature of’ some (‘severe COVID-19 disease’ and ‘death stage’)
However, these immune markers and profiles are also dependent on host qualities. Figure 5a shows that different qualities, such as biological sex (F/M), age, comorbidities, will infect disease outcomes. Figure 5b provides an CIDO representation of gene expression patterns in SARS-CoV-2 infected patients. Here we focus on the sex comparisons an example to illustrate the effect of biological sex to the disease outcome.
Increasing evidence show that male sex is a risk factor for a more severe COVID-19 disease outcome [12]. In one of the early studies with data in Wuhan, China, of 86 male COVID-19 patients, 12.8% (11/86) died; in comparison, of 82 female patients, 7.3% (6/82) died [34]. A cohort study of 17 million COVID-19 adult patients in England reported a strong association between male sex and risk of death [35]. Globally, approximately 60% of COVID-19 associated deaths are reported in men [36].
In CIDO, we represent the high susceptibility of male to the death using the following axiom:
‘male infected with SARS-CoV-2’: ‘has increased susceptibility compared to female to’ some ‘death stage’
This raises important question on the underlying molecular mechanisms underlying this sex difference and prompted further investigation using secondary analysis from the ImmPort studies. A total of 11 genes from Takashi et al. [12] were collected and compared for age and Body Mass Index corrected differences between patients and health care workers for each sex and is shown in Table 2. From this gene list, males and females showed statistically significant increases in 7 and 10 genes, respectively. To represent these differences between individuals (sex, exposure), we added new CIDO terms to distinguish between these differences as illustrated below (Fig. 6a).
‘symptomatic human male infected by SARS-CoV-2’: ‘organism susceptibly has up-regulated gene’ some ‘CCL4’
Such modeling allows us to perform semantic query as exemplified in Fig. 6c. In this example, we used a DL query to easily identify the number of up-regulated genes that are shared by male and female patients with COVID-19 (Fig. 6b).
Sex differences in gene ontology and DL query. a CIDO ontological representation of sex-based immune response for SARS-CoV-2. The genes listed are chosen from the results in Table 2. b DL Query infers properties provide a list to identify shared genes between males and females
In addition to the gene list identification and modeling, we further performed a secondary data analysis on the pathways. These gene lists were placed into Reactome to generate a set of pathways they were enriched. From these pathways, we restricted the background to 58 cytokines mapped from Takashi et al. [12] (out of 61 assays) and found that IL-10 immune pathways in both males and females were shown to be significant (p value of 8.47E-5 and 3.56E-5, respectively) despite differences in genes. The implication of such result is described in the following Discussion section.
Discussion
We have provided multiple contributions to the community with this effort. First, we demonstrated how we can apply a recursive XOD strategy to improve the CIDO by incorporating new knowledge learned from 6 publications from 6 ImmPort Studies related to COVID-19. Secondly, we demonstrate our better understanding of host responses to COVID-19, including sex differences, inflammatory cytokine responses, etc. Third, we performed secondary data analysis on the sex differences in response to COVID-19 infection. Our work showed that pathway analysis could provide more information than the gene list studies. Lastly, we provide a DL query to demonstrate how the integrated work can provide reasoning and inferences. We showed that our ontology framework supports knowledge representation, data and metadata standardization, and information query and analysis.
Our study presents a feasible strategy for new ontology updates by actively and progressively identifying new knowledge from literature mining and manual annotation of papers, or from our secondary data analysis of deposited data such as ImmPort. This is a process we need to do recursively. With each recursion, we reinforce the ontology coverage and quality. The basic XOD strategy informs how the CIDO can be developed, but it does not specifically inform how CIDO can be updated. For COVID-19 studies, expansive knowledge has been introduced by the literature and databases in a short period of time. Therefore, representing such data using CIDO has become challenging for our CIDO development team. Here we practice the XOD strategy and show that recursive XOD processes can help update the ontology. Instead of adopting a minimal updating principle for rapidly changing data, we used an active update pipeline. The information from other databases may also be incorporated.
New scientific insights about COVID-19 were also identified from the literature, summarized, and integrated into CIDO, leading to possible reasoning. For example, we were able to merge the epitope knowledge with the virus hierarchy, and virus variant information. It has been found that that many humans can develop protection against COVID-19 infection even when they have not been exposed to the SARS-CoV-2 virus, and such a phenomenon was owing to their protection to exposure to a common cold coronavirus and the cross reactivity of the epitopes between SARS-CoV-2 and some common cold coronaviruses [20, 26]. The viral variants also demonstrate the differences in COVID-19 transmissibility, vaccine efficacy and treatment efficacy [37, 38]. These findings have been semantically recorded in our CIDO knowledge representation.
Biomarkers and qualities (such as sex) in COVID-19 have been shown to manifest in negative disease outcomes (hospitalization, ICU, and mortality) and in differences in immune responses [11]. For example, age as an influence on comorbidities as a covariate for fatality is strongly sex specific [34]. Our secondary analysis using Reactome was inconclusive in most immune pathways; however, our Reactome study showed that both male and female patient populations had enriched expression of the IL-10 signaling pathway. The enrichment of the IL-10 signaling pathway is a novel finding from our secondary analysis of the ImmPort data since it was not reported in the original paper [12]. This prompted the next step of the recursive XOD to incorporate this new knowledge by looking to incorporate new terms and axioms related to IL-10. A recent March paper has proposed a hypothesis that IL-10 up-regulation is responsible for COVID-19 uniqueness in comparison to other coronaviruses [39]. SARS differs from COVID-19 in that IL-10 is only increased in convalescent SARS-CoV patients and but not with other SARS disease phenotypes [40, 41]. The IL-10 signaling pathway has many effects, and through direct macrophage and monocyte cells the pathway affects T-cell development and differentiation while enhancing B cell immune response [42]. Such mechanism appears to exist in both genders of patients, suggesting that it is a sex-independent mechanism. The authors also acknowledge that our finding is limited by the short list for background enrichment; however, multiple other papers have consistently found differences in outcomes [43, 44].
Despite strong evidence showing differences between sex, many countries have disaggregated COVID-19 data to sex which would occlude relations caused by differences in sex [45]. The modeling of different dispositions occurring due to differences in sex is able to help specify these tends which are not otherwise modeled in other ontologies.
The emergence of these new strains, such as B.1.1.7 and B.1.167+ [46] represent an important driver to our continuous application of the recursive XOD strategy for the future CIDO development. The modeling of characteristics of disease severity from their biomarkers and demographic differences are important additions to CIDO. Further incorporation of the transcriptome can be used to identify further molecular mechanisms and help elucidate further interactions in SARS-CoV-2.
As a formal ontology, CIDO is a logic computer-interpretable way of coronavirus-related knowledge representation, which supports reasoning and inference. CIDO does not represent specific instance-level data such as the data in the ImmPort database. Instead, CIDO represents coronavirus-related knowledge learned from the peer-reviewed publications or collected in well-annotated databases. On the other hand, ontology can support database data representation by ontologically representing the metadata or specific knowledge. For example, our group also maintains and develops the Vaccine Ontology (VO) [47, 48], which has been adopted and used in the ImmPort vaccine data annotation. This project is currently funded by an ImmPort-related NIH grant. Our ontology modeling outcomes are being shared with ImmPort, and we look forward to collaborating with the ImmPort team on how possibly our new CIDO knowledge representation can support their data storage and modeling.
Conclusion
ImmPort COVID-19 studies were further analyzed, and the knowledge learned were modeled and represent in the CIDO ontology. A recursive XOD strategy was proposed to systematically add the new knowledge to CIDO. New use cases include COVID-19 strain representation, the epitope cross-reactivity information and showing sex comparisons in responses to COVID-19 infection. Our use case studies demonstrate how we can actively and recursively update CIDO without suffering logical misinformation. Based on existing CIDO representation of various coronaviruses and proteins, we were able to quickly add the new knowledge of shared immune epitopes in different proteins of SARS-CoV-2 and other human coronaviruses that cause common colds.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and available as part of ImmPort or at https://github.com/CIDO-ontology/cido.
Abbreviations
- BFO:
-
Basic Formal Ontology
- ChEBI:
-
Chemical Entities of Biological Interest
- CIDO:
-
Coronavirus Infectious Disease Ontology
- DL query:
-
Description Logics query
- DOID:
-
Disease Ontology
- DRON:
-
Drug Ontology
- GISAID:
-
Global Initiative on Sharing Avian Influenza Data
- GO:
-
Gene Ontology
- IAO:
-
Information Artifact Ontology
- NCBITaxon:
-
NCBI organismal classification
- OBCS:
-
Ontology of Biological and Clinical Statistics
- OBI:
-
Ontology for Biomedical Investigations
- OBO:
-
The Open Biological and Biomedical Ontologies
- OWL:
-
Web Ontology Language
- PANGO:
-
Phylogenetic Assignment of Named Global Outbreak Lineages
- PATO:
-
Phenotypic Quality Ontology
- PR:
-
Protein Ontology
- RDF:
-
Resource Description Framework
- SPARQL:
-
SPARQL Protocol and RDF Query Language
- UBERON:
-
Uberon multi-species anatomy ontology
References
CDC COVID-19 data tracker weekly review [https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html].
Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5(1):180015. https://doi.org/10.1038/sdata.2018.15.
He Y, Yu H, Ong E, Wang Y, Liu Y, Huffman A, et al. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci Data. 2020;7(1):181. https://doi.org/10.1038/s41597-020-0523-6.
Ribeiro MM, Wassermann R, Flouris G, Antoniou G. Minimal change: relevance and recovery revisited. Artif Intell. 2013;201:59–80. https://doi.org/10.1016/j.artint.2013.06.001.
Solimando A, Guerrini G. Ontology adaptation upon updates. In: Extended semantic web conference. v. 7955. Springer; 2013. p. 34–45. https://link.springer.com/chapter/10.1007/978-3-642-41242-4_4.
Penaloza R, Thuluva AS. Iterative ontology updates using context labels. In: 1st workshop on belief change and non-monotonic reasoning in ontologies and databases. Association for Computer Linguistics: Buenos Aires; 2015.
He Y, Xiang Z, Zheng J, Lin Y, Overton JA, Ong E. The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability. J Biomed Semantics. 2018;9(1):3. https://doi.org/10.1186/s13326-017-0169-2.
Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–34. https://doi.org/10.1073/pnas.2003138117.
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e278. https://doi.org/10.1016/j.cell.2020.02.052.
Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5(1):237. https://doi.org/10.1038/s41392-020-00352-y.
Wolfe J, Safdar B, Madsen TE, Sethuraman KN, Becker B, Greenberg MR, et al. Sex- or gender-specific differences in the clinical presentation, outcome, and treatment of SARS-CoV-2. Clin Ther. 2021;43(3):557–71 e551. https://doi.org/10.1016/j.clinthera.2021.01.015.
Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588(7837):315–20. https://doi.org/10.1038/s41586-020-2700-3.
Arp R, Smith B, Spear AD. Building ontologies with basic formal ontology. Cambridge: MIT Press; 2015. https://doi.org/10.7551/mitpress/9780262527811.001.0001.
Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–5. https://doi.org/10.1038/nbt1346.
Ong E, Xiang Z, Zhao B, Liu Y, Lin Y, Zheng J, et al. Ontobee: a linked ontology data server to support ontology term dereferencing, linkage, query and integration. Nucleic Acids Res. 2017;45(D1):D347–52. https://doi.org/10.1093/nar/gkw918.
Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39(Web Server issue):W541–5.
Jupp S, Burdett T, Leroy C, Parkinson HE. A new Ontology Lookup Service at EMBL-EBI. In: SWAT4LS; 2015. p. 118–9.
Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. Jama. 2020;324(8):782–93. https://doi.org/10.1001/jama.2020.12839.
Gagliardi I, Patella G, Michael A, Serra R, Provenzano M, Andreucci M. COVID-19 and the kidney: from epidemiology to clinical practice. J Clin Med. 2020;9(8):2506.
Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. https://doi.org/10.1126/science.abd3871.
Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–43. https://doi.org/10.1093/nar/gky1006.
Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021. online ahead of publication.
Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. 2021;6:E335–45.
O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021. https://doi.org/10.1093/ve/veab064.
Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22(13):30494.
Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–501 e1415. https://doi.org/10.1016/j.cell.2020.05.015.
Liu Y, Chan W, Wang Z, Hur J, Xie J, Yu H, et al. Ontological and bioinformatic analysis of anti-coronavirus drugs and their implication for drug repurposing against COVID-19. Preprints. 2020;2020030413. https://www.preprints.org/manuscript/202003.0413/v1.
Liu Y, Hur J, Chan WKB, Wang Z, Xie J, Sun D, et al. Ontological modeling and analysis of experimentally or clinically verified drugs against coronavirus infection. Sci Data. 2021;8(1):16. https://doi.org/10.1038/s41597-021-00799-w.
Chung MK, Karnik S, Saef J, Bergmann C, Barnard J, Lederman MM, et al. SARS-CoV-2 and ACE2: the biology and clinical data settling the ARB and ACEI controversy. EBioMedicine. 2020;58:102907. https://doi.org/10.1016/j.ebiom.2020.102907.
Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–43. https://doi.org/10.1038/s41591-020-1051-9.
Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–9. https://doi.org/10.1038/s41586-020-2588-y.
Kang S, Tanaka T, Inoue H, Ono C, Hashimoto S, Kioi Y, et al. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc Natl Acad Sci U S A. 2020;117(36):22351–6. https://doi.org/10.1073/pnas.2010229117.
Jia R, Wang X, Liu P, Liang X, Ge Y, Tian H, et al. Mild cytokine elevation, moderate CD4(+) T cell response and abundant antibody production in children with COVID-19. Virol Sin. 2020;35(6):734–43. https://doi.org/10.1007/s12250-020-00265-8.
Meng Y, Wu P, Lu W, Liu K, Ma K, Huang L, et al. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: a retrospective study of 168 severe patients. PLoS Pathog. 2020;16(4):e1008520. https://doi.org/10.1371/journal.ppat.1008520.
Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584(7821):430–6. https://doi.org/10.1038/s41586-020-2521-4.
Gebhard C, Regitz-Zagrosek V, Neuhauser HK, Morgan R, Klein SL. Impact of sex and gender on COVID-19 outcomes in Europe. Biol Sex Differ. 2020;11(1):29. https://doi.org/10.1186/s13293-020-00304-9.
Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–5.
Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538).
Lindner HA, Velasquez SY, Thiel M, Kirschning T. Lung Protection vs. Infection Resolution: Interleukin 10 Suspected of Double-Dealing in COVID-19. Front Immunol. 2021;12:602130. https://doi.org/10.3389/fimmu.2021.602130.
Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11(6):715–22. https://doi.org/10.1111/j.1440-1843.2006.00942.x.
Zhang Y, Li J, Zhan Y, Wu L, Yu X, Zhang W, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72(8):4410–5. https://doi.org/10.1128/IAI.72.8.4410-4415.2004.
Moore KW, de Waal MR, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19(1):683–765. https://doi.org/10.1146/annurev.immunol.19.1.683.
Yu C, Littleton S, Giroux NS, Mathew R, Ding S, Kalnitsky J, et al. Mucosal associated invariant T (MAIT) cell responses differ by sex in COVID-19. Med (N Y). 2021;2:755–72.
Scully EP, Gupta A, Klein SL. Sex-biased clinical presentation and outcomes from COVID-19. Clin Microbiol Infect. 2021;27(8):1072–3. https://doi.org/10.1016/j.cmi.2021.03.027.
Hawkes S, Tanaka S, Pantazis A, Gautam A, Kiwuwa-Muyingo S, Buse K, et al. Recorded but not revealed: exploring the relationship between sex and gender, country income level, and COVID-19. Lancet Glob Health. 2021;9(6):e751–2. https://doi.org/10.1016/S2214-109X(21)00170-4.
Variants of concern or under investigation: data up to 28 April 2021 [https://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-cases-data]. Accessed 26 Apr.
Lin Y, He Y. Ontology representation and analysis of vaccine formulation and administration and their effects on vaccine immune responses. J Biomed Semantics. 2012;3(1):17. https://doi.org/10.1186/2041-1480-3-17.
Ozgur A, Xiang Z, Radev DR, He Y. Mining of vaccine-associated IFN-gamma gene interaction networks using the vaccine ontology. J Biomed Semantics. 2011;2(Suppl 2):S8.
Meng J, Xiao G, Zhang J, He X, Ou M, Bi J, et al. Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg Microbes Infect. 2020;9(1):757–60. https://doi.org/10.1080/22221751.2020.1746200.
Acknowledgements
We acknowledge the wide discussions and feedback from the experts in the host-microbiome interaction community and the biomedical ontology community.
Funding
This project was supported by the NIH grant 1UH2AI132931. It was also supported by a fund from the Michigan Medicine –Peking University Health Sciences Center Joint Institute for Clinical and Translational Research.
Author information
Authors and Affiliations
Contributions
AH: Ontology development, secondary data analysis, project design. AMM: Ontology development, immunology domain expert. JZ: Ontology development. NS and TB: Statistical data analysis. GW: Reactome pathway expert, pathway analysis. YH: Ontology development, microbiology and immunology domain expert, project design. AH and YH prepared the first manuscript draft. All contributed to manuscript preparation, discussion, and result interpretation. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Submitted to: Journal of Biomedical Semantics ICBO2021 series
https://www.biomedcentral.com/collections/icboseries
https://icbo2021.inf.unibz.it/call-for-papers/
https://docs.google.com/document/d/1pKx9hPXAJhMGfdevIic_BUAkHWBZwZpswPvMVcOtp4Q/edit
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huffman, A., Masci, A.M., Zheng, J. et al. CIDO ontology updates and secondary analysis of host responses to COVID-19 infection based on ImmPort reports and literature. J Biomed Semant 12, 18 (2021). https://doi.org/10.1186/s13326-021-00250-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13326-021-00250-4