Abstract
In recent years computational methods for molecular modeling have become a prime focus of computational biology and cheminformatics. Many dedicated systems exist for modeling specific classes of molecules such as proteins or small drug-like ligands. These are often heavily tailored toward the automated generation of molecular structures based on some meta-input by the user and are not intended for expert-driven structure assembly. Dedicated manual or semi-automated assembly software tools exist for a variety of molecule classes but are limited in the scope of structures they can produce. In this work we present BuildAMol, a highly flexible and extendable, general-purpose fragment-based molecular assembly toolkit. Written in Python and featuring a well-documented, user-friendly API, BuildAMol empowers researchers with a framework for detailed manual or semi-automated construction of diverse molecular models. Unlike specialized software, BuildAMol caters to a broad range of applications. We demonstrate its versatility across various use cases, encompassing generating metal complexes or the modeling of dendrimers or integrated into a drug discovery pipeline. By providing a robust foundation for expert-driven model building, BuildAMol holds promise as a valuable tool for the continuous integration and advancement of powerful deep learning techniques.
Scientific contribution
BuildAMol introduces a cutting-edge framework for molecular modeling that seamlessly blends versatility with user-friendly accessibility. This innovative toolkit integrates modeling, modification, optimization, and visualization functions within a unified API, and facilitates collaboration with other cheminformatics libraries. BuildAMol, with its shallow learning curve, serves as a versatile tool for various molecular applications while also laying the groundwork for the development of specialized software tools, contributing to the progress of molecular research and innovation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
The field of computational modeling for molecular structures has undergone a period of rapid expansion, driven by its growing importance in material sciences, pharmacology, and life sciences. This surge has led to the development of a diverse range of software solutions aimed at tackling various challenges in these fields. Furthermore, the continuous integration and advancement of powerful deep learning techniques within this domain promises to significantly enhance the impact of in silico structural modeling on future research applications.
Simplified molecular-input line-entry system (SMILES) [1] descriptors are a widely used and dominant method for generating three dimensional (3D) atomic structures of molecules. Their popularity stems from their efficiency and text-based format, making them well-suited for various applications. Consequently, many deep learning-based modeling tools heavily rely on SMILES strings as both input and output formats. Software libraries like RDKit [2] and Openbabel [3] effectively process SMILES to build molecular structures. However, this prevalent “all-at-once” approach, where the entire structure is constructed from a single SMILES string, presents limitations. For complex molecules like polymers, it can become computationally expensive. Additionally, limited user control over the final structure restricts its adaptation to specific research questions.
Fragment-based methods offer an alternative, by assembling larger structures incrementally from smaller, pre-defined components. This approach has gained traction in automated de novo molecule design frameworks like FRAME [4] and MolSLEPA [5], which focus on ligand design and material sciences, respectively. Beyond automated frameworks, fragment-based assembly empowers expert-driven, manual molecule creation. Users have control at each fragment addition, ensuring the resulting structure aligns with specific requirements at any intermediate stage of the building process. This approach not only be useful for manual design but also for refining predictions from deep learning models by providing more control over the final structure.
While popular libraries like RDKit can handle fragment assembly, they lack dedicated workflows for this purpose. Some libraries are adept at the assembly of new molecules from smaller components, such as the widely used Python-based Stk [6] or the Julia-based MolecularGraph.jl [7]. Especially Stk stands as a prominent example as its primary objective is to offer a comprehensive API for fragment-based assembly. However, its focus lies in material sciences, offering a powerful and generalizable but still somewhat domain-specific interface. Other Python libraries like mBuild [8] cater to specific software ecosystems like MoSDeF [9] for molecular simulations. While tools like Pygen-Structures [10] and Glycosylator [11] excel in constructing molecules from specialized data formats (CHARMM files) or specific molecule classes (glycans), respectively. Although these tools provide valuable functions, they are typically specialized and not easily adaptable for tasks beyond their original scope.
Here, we introduce BuildAMol, a general-purpose Python library designed for fragment-based molecular modeling. BuildAMol is designed to work on consumer-grade machines and can be used to manually and semi-automatically assemble models for diverse and complex molecular structures. BuildAMol is not restricted to specific molecule classes or limited by pre-defined chemical reasoning but offers complete user freedom in assembling fragments, akin to the familiar analog molecule building kits used by chemists. Furthermore, BuildAMol’s functionalities extend beyond de novo assembly. It enables modification and optimization of existing structures, conformational sampling, generation of customizable and interactive visualizations, and seamless integration with other cheminformatics libraries. This comprehensive suite of tools fosters a streamlined workflow for diverse applications. BuildAMol prioritizes user-friendliness through a streamlined interface, minimizing manual input while maximizing user control over the assembly process.
Implementation
BuildAMol is designed for modularity and ease of use. The six primary sub-packages of BuildAMol are: core, graphs, optimizers, resources, structural, and utils, that are automatically loaded upon importing the main BuildAMol library. An optional seventh sub-package, (extensions), provides specialized functionalities and needs to be imported separately (Fig. 1).
Software overview. A Graphical summary of BuildAMol, its seven packages, and their functional interplay. B Integrations with external libraries and supported file formats. C Overview of built-in reference data. D Overview of supported molecular visualizations. All panels show the small alternative reading frame peptide SPRWWPTCL of Intestinal carboxyl esterase found in Renal cell carcinoma [12]
Software overview
Core
The core sub-package of BuildAMol provides the essential framework for constructing molecular structures. It provides foundational classes representing the building blocks of molecules (Atom and Residue), as well as higher-level classes (Molecule and Linkage) to handle the assembly process. BuildAMol leverages a hierarchical data structure, similar to the one used by Biopython [13]. In this approach, complex molecules are organized as a series of nested building blocks. For instance, individual Atom objects are grouped into a Residue object representing a molecular fragment. This hierarchical organization allows for efficient access and manipulation of specific parts within complex molecules, a significant advantage for fragment-based assembly workflows.
BuildAMol simplifies user interaction by channeling most operations through the Molecule class. This class acts as a central hub, providing methods for users to perform various actions on molecular structures. For convenience, some frequently used functions, like reading PDB files or connecting fragments, are available as both standalone functions and methods within the Molecule class. The key distinction lies in their behavior: standalone functions typically create a new copy of the molecule, whereas methods directly modify the existing molecule object. However, both functionalities can be customized using optional arguments.
The Linkage class plays a crucial role in fragment assembly workflows. It defines how atoms from different molecules should be connected during the building process. Additionally, the package offers convenient top-level functions like acetylate or carboxylate to add specific functional groups to a molecule at defined positions.
Furthermore, BuildAMol introduces a unique operator-based syntax called “Molecular Arithmetics.” This user-friendly syntax allows intuitive molecule assembly: addition (+) facilitates combining fragments, subtraction (-) enables the removal of substructures like atoms or residues, and multiplication (*) allows for the construction of regular polymers, offering a valuable short-hand notation to accelerate the modeling for users who do not wish or require more elaborate customization in their assembly workflow.
Graphs
BuildAMol implements molecular connectivity graphs using NetworkX [14]. Graphs are available at the atom level and as an abstraction at the residue level, primarily for visualization or low-resolution optimization. Beyond storing a molecule’s connectivity, these graphs serve as inputs for BuildAMol’s in-house optimization suite (Optimizers sub-package). Users typically will not interact directly with the graphs package. Instead, they can work with graph objects exported from the Molecule class using methods such as Molecule.get_atom_graph or Molecule.get_residue_graph for conformational optimization.
Optimizers
BuildAMol provides several methods to perform conformational optimization on a target molecule. If the RDKit library is available, BuildAMol can leverage its forcefield-based optimization to automatically obtain an energy-minimized conformation. BuildAMol provides its own customizable optimization environment built using OpenAI Gym [15]. Various algorithms implemented in NumPy [16], including genetic algorithms, particle swarm optimization, simulated annealing, and Scipy-based stochastic gradient descent are available. In addition to the default setup provided by the Molecule.optimize method and the top-level optimize, the user can create custom optimization environments.
Resources
The built-in reference data for chemical compounds relevant to fragment-based assembly are stored in the Resources sub-package. This sub-package provides numerous functions to query compounds, load existing data or read new data from files, add molecules to the reference dataset, as well as save datasets to files. The default reference data is a subset of the Protein-Data-Bank’s Component Dictionary (PDBCD), containing a chosen selection of 7630 molecules out of the nearly 40,000 available. To facilitate efficient access, the reference data is further categorized into five individually loadable datasets: small_molecules, amino_acids, nucleotides, sugars, and lipids. Additionally, users can load their own custom datasets in the mmCIF format for further flexibility. Beyond fragment data, we also included a selection of molecular linkages derived from CHARMM topology files (Patches). Custom linkage defined in the CHARMM topology format can also be imported into the reference dataset. Figure 1C provides a graphical summary of the built-in reference data. For user convenience, all core functionalities of the Resources sub-package are automatically imported upon loading the main BuildAMol library. This eliminates the need for manual interaction with the sub-package itself in most use cases.
Structural
The Structural sub-package forms the core of BuildAMol’s structural manipulation capabilities. It encompasses functionalities related to atom-level manipulations, including algorithms used to compute atomic placements during fragment assembly. This sub-package provides a rich set of functions and data classes for internal use, such as fundamental mathematical operations on vectors and matrices, explicit computations on molecular structures, and inference tasks like adding missing hydrogen atoms. The Structural sub-package serves as a foundation for most other BuildAMol functionalities. Many of its functionalities are also directly accessible through methods of the Molecule class. Therefore, in most use cases, users will not need to interact directly with the Structural sub-package itself.
Utils
The Utils sub-package provides a collection of utility functions and helpers that support BuildAMol’s internal operations. This includes managing optional dependencies, general constants, and the core functionalities for molecular visualization.
BuildAMol’s built-in visualization leverages Plotly [17] to generate interactive 3D representations of molecular structures. These visualizations can be customized with points, lines, annotations, and hover data to highlight specific features or regions of interest. While Plotly is the default option, BuildAMol offers additional visualization support using RDKit (2D only), Py3DMol [18], and NGLView [19], provided they are installed (Fig. 1D). All visualization functionalities are conveniently accessible through methods of the Molecule class, minimizing the need for direct interaction with the Utils sub-package itself.
Extensions
The Extensions sub-package serves as an open hub for expanding BuildAMol’s functionalities. It currently provides dedicated implementations for specific modeling problems. Currently, we implemented packages to generate linear and cyclic polycarbons, nanotubes, metal complexes, rotaxanes, glycans, small peptides, fatty acids, mono-, di-, and triacylglycerols, as well as phospho- and sphingolipids.
Connecting molecules
BuildAMol leverages an atom-substitution strategy to connect molecular fragments, enabling the direct spatial positioning of one molecule relative to another. This approach involves designating a set of four atoms, two from each fragment. A rigid body transformation is then employed to precisely align the fragments by superimposing two atoms from one fragment onto their counterparts in the other fragment. This ensures proper spatial arrangement for bonding. Subsequently, a new bond is formed between a designated atom from each fragment, while the corresponding atoms used for alignment are removed. This method ensures the resulting molecule has realistic bond lengths and angles, provided both fragment molecules are chemically sound.
While BuildAMol is able to generate a wide variety of molecular structures, its atom-substitution strategy introduces limitations for certain reaction types. Since it requires an atomic “leaving group” for substitution, BuildAMol cannot directly simulate reactions that do not involve atom removal, such as the reduction of double bonds. Additionally, the system is primarily designed to create linear or branched structures. Although optimization techniques can enable BuildAMol to generate cyclic molecules, it is not the most efficient approach for this specific task.
Two modes of fragment assembly
Superimposing fragments based on a single reference bond offers a fast and convenient alignment method. However, with only two points of reference, the relative orientation of the fragments remains undefined, potentially leading to sub-optimal conformations. BuildAMol addresses this issue by providing two options. The first approach requires defining a detailed Linkage. This linkage specifies the internal coordinates, such as bond lengths, angles, and dihedrals, for the atoms surrounding the newly formed bond. This additional geometric information allows BuildAMol to place the fragment precisely in the desired orientation. However, defining such detailed geometry might not always be feasible or necessary. Therefore, by default, BuildAMol will perform a small-scale optimization around the new bond in order to rotate the incoming fragment into a chemically sound orientation.
In practice, the mode of assembly is automatically determined by BuildAMol behind the scenes. To connect molecules in BuildAMol the user may use the Molecule.attach method or the top-level connect function. Alternatively, the user may use Molecular Arithmetics to “add” molecules together using the + operator. A more detailed description of the assembly modes is provided in the supplementary materials.
Chemical reactions
While BuildAMol was not designed to simulate true chemical reactions, it offers functionalities to achieve similar outcomes for specific reaction types. This is achieved by a user-extensible library of functional groups. When reacting two fragments, functional groups automatically infer the binder and deleter atoms needed to link both fragments. Therefore, substitution reactions can be imitated to a certain extent, by using either the Molecule.react_with method or top-level react function or the + operator. It is important to note that due to its core principle of atom substitution, BuildAMol currently cannot handle addition reactions, where a new bond is formed without the removal of existing atoms.
The optimization suite
Conformational optimization
BuildAMol implements a torsional optimization scheme to improve molecular conformations. By rotating a specific part of the molecule around a chosen bond’s axis, a new conformation is generated. This approach inherently avoids introducing invalid bond lengths or angles, assuming the initial structure is valid. Also, compared to traditional “translational” optimization, this “torsional” approach typically explores a smaller search space. This is because the number of bonds in a molecule is usually less than, and can never exceed, the number of atoms. Additionally, the search space can be further reduced by strategically subsetting the bonds considered during optimization.
We implemented a basic OpenAI Gym environment named the Rotatron that accepts a molecular graph (either at the atom or residue level) and an optional list of rotatable edges to optimize. If not provided, rotatable edges are inferred directly from the graph. Using the Rotatron as a parent class, we implemented three optimization environments that use different heuristics to evaluate a given conformation. (1) The DistanceRotatron, which serves as the default optimization environment in BuildAMol. It aims to maximize pairwise distances between graph nodes in order to obtain a conformation with maximal spatial occupancy. Since the heuristic computes distances between pairs of nodes, its computational load can be heavy for large input graphs. To address this, we developed (2) the OverlapRotatron. This environment models all nodes between two rotatable edges, a so-called “rotation unit”, using a Gaussian Mixture Model. The environment’s heuristic is set toward minimizing the overlap between all Gaussians in order to achieve a conformation with maximal spatial occupancy. Both the DistanceRotatron and OverlapRotatron strictly perform “unfolding” operations and are not suited for optimizing molecules with prominent non-covalent interactions such as Hydrogen bonds. To account for cases where such purely geometric considerations may not be sufficient for optimization, we also developed (3) the ForceFieldRotatron. This environment uses RDKit’s Merck Molecular Force Field (MMFF) to compute the molecular energy of a particular conformation, which directly serves as the optimization metric.
Circularization
At its core, BuildAMol was designed to create linear or branched molecules rather than circular structures. To create circular molecules, BuildAMol provides an extended version of the Rotatron, named the Circulatron. This environment works on a linear molecule graph alongside instructions on ultimately circularizing the structure. The Circulatron then uses one of the three above-mentioned environments to keep track of the quality of generated conformations while trying to superimpose binder and deleter atoms that must be provided during initialization.
Spatial optimization
BuildAMol can also be used to construct multi-molecule systems that require a molecule’s specific placement and orientation in three-dimensional space. While methods such as Molecule.move_to, Molecule.rotate, or Molecule.align_to are designed to facilitate the manual arrangement of a molecule in space, it can be difficult for a user to identify the right location and orientation a priori. Thus, to facilitate the placement of molecules in a particular system, we developed an optimization environment called the Translatron. This environment optimizes a translation and rotation vector along all three primary spatial axes, which are applied to the entire molecule. Consequently, this environment will not alter the molecule’s conformation but only its global placement and orientation. To guide the optimization process, the environment requires the user to provide a constraint function that returns a metric the environment tries to minimize. Thus, the user must define a suitable heuristic function that describes the placement problem they want to solve.
User-guided optimization
While the three primary conformational optimization environments employ a fixed heuristic that is not customizable by the user, situations like those encountered in the Circulatron may necessitate special considerations during optimization. To address this, we introduced a more versatile ConstraintRotatron class. This class acts as a wrapper, utilizing one of the three main environments to assess the created conformations. It also requires a user-provided constraint function to enhance the heuristic as per \(score = eval(s) + constraint(s)\), where eval is the evaluation function of any optimization environment and s denotes the current state that is evaluated. In this way, users may direct the optimization process by their specific needs.
Optimization algorithms
We implemented several classical optimization algorithms directly in BuildAMol, including a genetic algorithm (GA), a global-best particle swarm optimization (PSO), and simulated annealing. Additionally, molecules can be forwarded directly to SciPy’s minimize optimization suite, where stochastic gradient descent (SGD) is used by default, though any algorithm available within the suite can be utilized.
Performance enhancements
Numba [20] is a Just-In-Time Compiler (JIT) to improve the runtime efficiency of Python code and primarily works with Numpy. We implemented several functions of the optimization environment setup and optimization algorithms as standard Numpy and Numba versions to allow for a greater speed-up. BuildAMol also offers parallel computing using Python’s built-in multiprocessing library to enhance performance for certain functions.
Results
Showcase of example molecules
To evaluate the performance of BuildAMol, we generated three different benchmark. We measured the runtime after imports as well as the number of lines of code. The lines of code were counted after formatting with the Black formatter [21] and include import-lines but typically exclude intermediary visualizations added purely for clarity in the tutorial code. The code for all examples is available in the BuildAMol documentation on ReadTheDocs.
To demonstrate BuildAMol’s capability of building a wide range of molecules with concise code, we constructed several structures from various molecular classes and sizes (Fig. 2). This included a small rotaxane [22], a glycan, a circular poly-Histidine, and two dendrimers [23, 24].
The rotaxane was built using three fragments for the axle and one single fragment for the ring. To align the ring around the axle molecule, we wrote a RotaxaneBuilder class, which is available as part of the Extensions. The complete example comprises 28 lines of code.
For the glycan model, all necessary fragments and linkages are available in the built-in reference dataset, and the complete example comprises 11 lines of code.
The poly-histidine was initially created as a linear peptide, which was then pseudo-circularized by connecting the first and last residues. This unrealistic conformation was optimized using RDKit’s optimization suite to obtain the final circular structure. The code comprises 8 lines of code.
The polyphenylene dendrimer was modeled using only a single fragment molecule (benzene) and required 20 lines of code.
Finally, the open-resorcinarene dendrimer was built from “inside” to “outside”, by incrementally attaching new fragments to multiple residues and optimizing structural intermediaries. The final code comprises 43 lines.
Comparing to Stk
To evaluate BuildAMol’s performance and capabilities, we compared it to the Stk library, the closest existing software for molecular construction. We focused on five diverse examples from the Stk documentation with publicly available source code (Fig. 3). These examples encompassed a wide range of molecular structures: (1) an aminated cycloheptane ring, (2) a macrocycle of butylamine molecules, (3) a metal complex, (4) a linear polymer, and (5) a simple rotaxane with three cycles around a linear axle.
Our evaluation included a comparison of code conciseness and runtime performance between BuildAMol and Stk. The hardware used for this comparison was an octa-core Intel Core i9 processor with 2.3 GHz clock speed and 32 GB of RAM.
In every instance, using BuildAMol required fewer lines of code to accomplish identical molecular construction, typically resulting in a 50% decrease compared to Stk code (Fig. 3, upper bar chart). While BuildAMol demonstrated faster execution times in examples 1, 2, and 4 (Fig. 3, lower bar chart), for example 5 (a rotaxane), the execution time was comparable for both. The only example where Stk clearly outperformed BuildAMol was example 3, the metal complex. This can be attributed to BuildAMol’s sequential optimizations for ligand placement around the metal center. This approach is computationally expensive and can potentially lead to unstable structures. In contrast, Stk utilizes a dedicated geometric approach for metal complex generation, resulting in faster and more stable performance. It is important to note, however, that the Stk-generated metal complex exhibited misalignment in half of its Nitrogen atoms.
Since the Stk core library does not consider the chemical validity of the generated structures and relies on the external library Stko for structure optimization Stko, we compared BuildAMol’s outputs to both the raw and optimized structures from Stk or Stk+Stko. In each case we optimized Stk-raw structures using Stko’s UFF Force Field implementation. In most cases Stk+Stko structure outputs were on par with those of BuildAMol. In case of the metal complex (3) the Nitrogen atoms were properly aligned but at the cost of bond distortions in the ligands. In the case of the rotaxane (5), after optimization with Stko, the axle molecule’s distorted bonds were fixed, but the cycles were colliding with the main axle.
Although BuildAMol is capable of generating models for all molecule classes supported by Stk, including caged structures and macro-frameworks, it is important to note that, currently, BuildAMol does not incorporate specific methods tailored toward modeling these structures.
Benchmarking optimization. A A graphical summary of the benchmarking workflow. We optimized the AtomGraph and ResidueGraph of the input molecule 20 times in parallel with PSO using all three optimization environments, respectively. All optimizations were done on default settings. B The number of clashes in the final conformations are shown as mode (upper) and mean + standard deviation (lower). C A more detailed view of the distributions of clashes in final conformations split by graph and environment. D The total measured computation times (mean + standard deviation) for environment setup and optimization. The code was run on an octa-core Intel Core i9 processor with 2.3GHz and 32 Gigabyte of RAM. E The test molecule used for comparison between BuildAMol, Ginger, and Frog2. The 3D view shows the UFF-minimized conformation. F Upper bar chart: UFF-based molecular energy of generated conformers. A black dashed line denotes the energy of the UFF-minimized conformer. Lower bar chart: RMSD of generated conformers compared to the UFF-minimized conformation. Shown is the mean + standard deviation. G Visual overlay of all generated conformers. The coloring matches the bar charts in F
Benchmarking optimization
We evaluated the performance of different optimization environments on a highly branched dendrimer, composed of 657 atoms in total (Fig. 4). We optimized the atom and residue graphs on a random subset of 15 bonds using each optimization environment and global-best particle swarm optimization. We performed 20 parallel optimizations in each case and repeated the entire workflow 20 times, resulting in a total of 400 generated conformations for each combination of graph and environment (Fig. 4A).
Our results indicate that, even without hyperparameter tuning of the environments, BuildAMol produced clash-free conformations reliably in almost all cases. The mode of clashes in optimized conformations was zero for all combinations of graph and environment except for the ResidueGraph+ForceFieldRotatron and ResidueGraph+OverlapRotatron (Fig. 4B). Inspection of the distributions of clashes in the final conformations shows that the AtomGraph+DistanceRotatron and AtomGraph+ForceFieldRotatron markedly tend toward producing clash-free structures, while any combination with the OverlapRotatron yields poorer results due to the greater structural abstraction (Fig. 4C).
Noticeably poor performance is exhibited by theResidueGraph+ForceFieldRotatron, where the worst conformation comprises 40 clashes.This lack of performance can be explained by the graph input. While the environment computes molecular energy, which should be a highly accurate metric for conformer evaluation, the input residue graph is not a valid chemical structure and, therefore, its molecular energy is a meaningless metric to optimize. Hence, we strongly discourage users from optimizing structures in this way.
While the DistanceRotatron showed the most consistent behavior, it was also by far the slowest environment to compute. Both the OverlapRotatron and ForceFieldRotatron were roughly on par in terms of computation time (Fig. 4D).
We also evaluated BuildAMol’s conformer generation capacities on a small drug-like molecule of 49 atoms (Fig. 4E). We generated 50 conformers with default optimization settings using the DistanceRotatron and automatic edge selection. For comparison, we generated an equivalent set of conformers using Ginger [25] and Frog2 [26]. Ginger uses a generative deep learning framework paired with a force-field minimization, while Frog2 utilizes a Monte Carlo sampling mechanism similar to BuildAMol.
We evaluated the generated conformers by computing their molecular energy with RDKit’s Universal Force Field (UFF). Additionally, we determined the atomic Root Mean Square Deviation (RMSD) between each generated conformer and the molecule’s UFF-minimized structure.
Since both BuildAMol and Frog2 employ a torsional optimization scheme, their performances were comparable. The generated conformations exhibited approximately double the energy of the UFF-minimized structure (Fig. 4F, upper bar chart). Since Ginger employs an energy minimization step, the generated conformers showed a lower energy level, similar to the UFF-minimized one. However, all three tools produced conformers with comparable RMSD values (Fig. 4F, lower bar chart). Visual inspection showed that Frog2 produced many similar conformers resulting in a clustered appearance (Fig. 4G), while both BuildAMol and Ginger showed greater diversity in the produced conformations. As expected, Ginger’s conformers closely resembled the UFF-minimized structure, while BuildAMol’s conformers tended to be elongated due to the DistanceRotatron optimization.
While BuildAMol demonstrated comparable performance to other conformer generation tools in our tests, the use of dedicated software remains advantageous for specific applications. For instance, small drug-like molecules can benefit from a wider range of specialized options. Moreover, BuildAMol’s current limitations preclude its effective application to macrocyclic molecules. In such cases, tools like OpenEye’s OMEGA [27] are recommended.
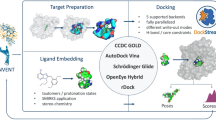
Extended use cases. A A metal-organic framework constructed from benzene and a metal complex. B Conformational sampling of a molecular compound. C Protein-ligand design. The upper panel shows the design workflow. Steps involving BuildAMol are highlighted with the BuildAMol logo. Lower left panel: our designed ligand (light blue) and the true ligand Risperidone (sand-colored) are shown overlaid on the dopamine D2 receptor. Lower right panel: modified versions of our ligand docked on the same protein
Extended use cases
While BuildAMol’s core functionality is the generation of molecular models from smaller fragments, we also explored more exotic use cases that go beyond “simple” fragment-based molecular assembly. Here, we present three different examples.
BuildAMol was primarily intended for solvated organic molecules that have conformational freedom. As such, modeling pseudo-crystalline structures was not our primary objective, especially in light of Stk which is able to handle such structures well. Nevertheless, we demonstrate that geometrically regular structures can be easily created using BuildAMol by employing methods such as align_to, move, and merge instead of attach which is conventionally used to assemble fragments. Here we demonstrate an example metal organic framework with a pillard paddlewheel structure [28] which we modeled from a metal complex fragment and benzene (Fig. 5A). The metal complex was generated with Stk.
In a second example, we used BuildAMol’s parallel optimization capacities to perform conformational sampling. We repeatedly selected a random subset of bonds within the target molecule and performed independent optimizations on them. Figure 5B shows an overlay of 50 thus sampled partial conformations.
Finally, in the third case we created a simple automated protein-ligand design pipeline using BuildAMol. To that end, we prepared a small library of some 200 fragments of small organic molecules. We then developed a class to assemble fragments based on instructions encoded numerically in Numpy arrays. The assembled molecules were then passed to the software library dockstring [29] to generate a docking score. For our example case, we chose the dopamine D2 receptor (Uniprot ID F8VUV1), which is bound by the drug Risperidone. Our “pipeline’s” main function would generate a molecule from a Numpy array and return a docking score for use with an optimization algorithm. For the example, we used Scipy’s Nelder-Mead algorithm which we ran for five iterations only. We repeated the test three times. Interestingly, our simple pipeline was already able to generate molecules that bind in the same pocket as Risperidone (Fig. 5C, lower left panel). Moreover, our best prediction (the blue ligand in Fig. 5C, lower left panel) showed some features that were structurally similar to Riesperidone and produced a docking score of − 11.6, which is in a similar range to Risperidone’s own docking score of − 11.9. As a proof of concept to demonstrate BuildAMol in a workflow involving deep learning applications, we performed a follow-up experiment where we created multiple derivatives from the generated ligand by adding various functional groups to one position. We then successfully docked the derivatives using the deep learning tool DiffDock [30], presenting a practical illustration of BuildAMol’s compatibility with state-of-the-art deep learning tools (Fig. 5C, lower right panel).
Conclusion
In this work, we presented BuildAMol, a versatile and user-friendly Python library designed to empower researchers in fragment-based molecular modeling. BuildAMol caters to a broad range of applications, from de novo assembly of complex molecules to the modification and optimization of existing structures. Its focus on user control and extensibility supports both manual and semi-automated workflows, making it suitable for expert-driven modeling tasks.
BuildAMol’s ability to handle diverse molecule classes and integrate with established cheminformatics libraries positions it as a valuable tool for various scientific pursuits. By prioritizing user-friendliness and offering a streamlined interface, BuildAMol minimizes manual input while maximizing control over the molecular assembly process. Coupled with its extensibility, BuildAMol is a promising platform for future advancements in fragment-based modeling, particularly when integrated with powerful deep-learning techniques. The continuous integration and advancement of these techniques within this domain promise to significantly enhance the impact of in silico structural modeling on future research initiatives.
Availability of data and materials
The source code of BuildAMol is available via GitHub at https://github.com/NoahHenrikKleinschmidt/buildamol. Comprehensive documentation with tutorials is also available on ReadTheDocs at https://biobuild.readthedocs.io. Code to recreate examples presented in this work is freely available via the documentation or GitHub repository.
References
Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci 28(1):31–36. https://doi.org/10.1021/ci00057a005
Landrum G, Tosco P, Kelley B, Ric, Cosgrove D, Sriniker, Gedeck, Vianello R, NadineSchneider, Kawashima END, Jones G, Dalke A, Cole B, Swain M, Turk S, AlexanderSavelyev, Vaucher A., Wójcikowski M, Take I, Probst D, Ujihara K, Scalfani VF, Godin, Lehtivarjo J, Pahl A, Walker R, Berenger F. jasondbiggs, strets123: Rdkit/rdkit: 2023_03_2 (Q1 2023) Release. https://doi.org/10.5281/zenodo.8053810
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: An open chemical toolbox. J Cheminform 3(1):1–14
Powers AS, Yu HH, Suriana P, Koodli RV, Lu T, Paggi JM, Dror RO (2023) Geometric deep learning for structure-based ligand design. ACS Central Sci 9(12):2257–2267. https://doi.org/10.1021/acscentsci.3c00572
Li J, Sumita M, Tamura R, Tsuda K (2023) Interpretable fragment-based molecule design with self-learning entropic population annealing. Adv Intell Syst 5(10):2300189. https://doi.org/10.1002/aisy.202300189
Turcani L, Tarzia A, Szczypiński FT, Jelfs KE (2021) stk: An extendable Python framework for automated molecular and supramolecular structure assembly and discovery. J Chem Phys 154(21):214102. https://doi.org/10.1063/5.0049708
Matsuoka S, Holy T, Hhaensel Henle A, Skim R, McGrath T, Box W (2024) mojaie/MolecularGraph.jl: v0.17.1 https://doi.org/10.5281/zenodo.12789286
Klein C, Sallai J, Jones TJ, Iacovella CR, McCabe C, Cummings PT (2016) A hierarchical, component based approach to screening properties of soft matter https://doi.org/10.1007/978-981-10-1128-3_5
Cummings PT, McCabe C, Iacovella CR, Ledeczi A, Jankowski E, Jayaraman A, Palmer JC, Maginn EJ, Glotzer SC, Anderson JA, Ilja Siepmann J, Potoff J, Matsumoto RA, Gilmer JB, DeFever RS, Singh R, Crawford B (2021) Open-source molecular modeling software in chemical engineering focusing on the molecular simulation design framework. AIChE J 67(3):17206. https://doi.org/10.1002/aic.17206
Hesketh T (2020) pygen-structures: A python package to generate 3d molecular structures for simulations using the charmm forcefield. J Open Source Softw 5(48):2157. https://doi.org/10.21105/joss.02157
Lemmin T, Soto C (2019) Glycosylator: a python framework for the rapid modeling of glycans. BMC Bioinform 20(1):513. https://doi.org/10.1186/s12859-019-3097-6
Ho O, Green WR (2006) Alternative translational products and cryptic t cell epitopes: expecting the unexpected. The Journal of Immunology 177(12):8283–8289
Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, Hoon MJL (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25(11):1422–1423. https://doi.org/10.1093/bioinformatics/btp163
Hagberg A, Swart P, S Chult D (2008) Exploring network structure, dynamics, and function using networkx. Technical report, Los Alamos National Lab.(LANL), Los Alamos, NM (United States)
Brockman G, Cheung V, Pettersson L, Schneider J, Schulman J, Tang J, Zaremba W (2016) OpenAI Gym. cite arxiv:1606.01540
Harris CR, Millman KJ, Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, Kerkwijk MH, Brett M, Haldane A, Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE (2020) Array programming with NumPy. Nature 585(7825):357–362. https://doi.org/10.1038/s41586-020-2649-2
Inc., P.T.: Collaborative Data Science. https://plot.ly
Rego N, Koes D (2014) 3Dmol.js: molecular visualization with WebGL. Bioinformatics 31(8):1322–1324. https://doi.org/10.1093/bioinformatics/btu829
Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlić A, Rose PW (2018) NGL viewer: web-based molecular graphics for large complexes. Bioinformatics 34(21):3755–3758. https://doi.org/10.1093/bioinformatics/bty419
Lam SK, Pitrou A, Seibert S (2015) Numba: A llvm-based python jit compiler. In: Proceedings of the Second Workshop on the LLVM Compiler Infrastructure in HPC, pp. 1–6
Langa, L., contributors to Black: Black: The Uncompromising Python Code Formatter. https://github.com/psf/black
Tian C, Fielden SDP, Whitehead GFS, Vitorica-Yrezabal IJ, Leigh DA (2020) Weak functional group interactions revealed through metal-free active template rotaxane synthesis. Nat Commun 11(1):744. https://doi.org/10.1038/s41467-020-14576-7
Bauer RE, Enkelmann V, Wiesler UM, Berresheim AJ, Müllen K (2002) Single-crystal structures of polyphenylene dendrimers. Chem Eur J 8(17):3858–3864. https://doi.org/10.1002/1521-3765(20020902)8:17(3858::AID-CHEM3858)3.0.CO;2-5
Pedro-Hernandez DL, Martinez-Garcia M (2022) Synthesis of open-resorcinarene dendrimers with l-serine (ibuprofen) derivatives. Curr Org Chem 26(1):71–80
Raush E, Abagyan R, Totrov M (2024) Efficient generation of conformer ensembles using internal coordinates and a generative directional graph convolution neural network. J Chem Theory Comput 20(9):4054–4063. https://doi.org/10.1021/acs.jctc.4c00280
Miteva MA, Guyon F, Tuffery P (2010) Frog2: efficient 3D conformation ensemble generator for small compounds. Nucleic Acids Res 38(Web Server), 622–627 https://doi.org/10.1093/nar/gkq325
Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and Cambridge structural database. J Chem Inf Model 50(4):572–584. https://doi.org/10.1021/ci100031x
Deria P, Mondloch JE, Karagiaridi O, Bury W, Hupp JT, Farha OK (2014) Beyond post-synthesis modification: evolution of metal-organic frameworks via building block replacement. Chem Soc Rev 43:5896–5912. https://doi.org/10.1039/C4CS00067F
García-Ortegón M, Simm GNC, Tripp AJ, Hernández-Lobato JM, Bender A, Bacallado S (2022) Dockstring: easy molecular docking yields better benchmarks for ligand design. J Chem Inf Model 62(15):3486–3502. https://doi.org/10.1021/acs.jcim.1c01334
Corso G, Stärk H, Jing B, Barzilay R, Jaakkola T (2023) Diffdock: Diffusion steps, twists, and turns for molecular docking. In: International Conference on Learning Representations (ICLR)
Funding
This work has been supported by the Swiss National Science Foundation (SNSF: PCEFP3_194606).
Author information
Authors and Affiliations
Contributions
N.K. designed, implemented, and tested the BuildAMol toolkit. N.K. and T.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kleinschmidt, N., Lemmin, T. BuildAMol: a versatile Python toolkit for fragment-based molecular design. J Cheminform 16, 104 (2024). https://doi.org/10.1186/s13321-024-00900-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13321-024-00900-6