Abstract
Background
Many laboratories have described the in vitro isolation of multipotent cells with stem cell properties from the skin of various species termed skin-derived stem cells (SDSCs). However, the cellular origin of these cells and their capability to give rise, among various cell types, to male germ cells, remain largely unexplored.
Methods
SDSCs were isolated from newborn mice skin, and then differentiated into primordial germ cell-like cells (PGCLCs) in vitro. Single-cell RNA sequencing (scRNA-seq) was then applied to dissect the cellular origin of SDSCs using cells isolated from newborn mouse skin and SDSC colonies. Based on an optimized culture strategy, we successfully generated spermatogonial stem cell-like cells (SSCLCs) in vitro.
Results
Here, using scRNA-seq and analyzing the profile of 7543 single-cell transcriptomes from newborn mouse skin and SDSCs, we discovered that they mainly consist of multipotent papillary dermal fibroblast progenitors (pDFPs) residing in the dermal layer. Moreover, we found that epidermal growth factor (EGF) signaling is pivotal for the capability of these progenitors to proliferate and form large colonies in vitro. Finally, we optimized the protocol to efficiently generate PGCLCs from SDSCs. Furthermore, PGCLCs were induced into SSCLCs and these SSCLCs showed meiotic potential when cultured with testicular organoids.
Conclusions
Our findings here identify pDFPs as SDSCs derived from newborn skin and show for the first time that such precursors can be induced to generate cells of the male germline.
Similar content being viewed by others
Introduction
Over the past few decades, scientists have explored whether functional gametes can be obtained from many different types of stem cells under in vitro conditions [1, 2]. Recently, complete spermatogenesis and oogenesis under in vitro conditions have been established from mouse pluripotent embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) [3, 4]. As far as we know, it has not been possible to reproduce these results starting from adult stem cells (ASCs), although some types of ASCs were shown to be capable of generating germ cell-like cells (GCLCs) in vitro [5,6,7,8,9,10].
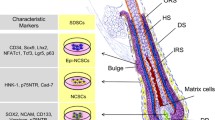
In 2001, skin-derived precursors representing novel multipotent adult stem cells, distinct from epidermal stem cells (EPSCs) and dermal mesenchymal stem cells (MSCs), were isolated from juvenile and adult mice [11]. These cells, possibly derived from neural crest or mesodermal cells, were reported to reside in the dermis and be able to generate cells of different lineages, such as glia, adipocytes, neuron, and smooth muscle cells. Cells with similar characteristics termed skin-derived stem cells (SDSCs), were isolated also from other species including pigs and humans [12,13,14]. In 2006, Dyce and colleagues reported the generation of oocyte-like cells (OLCs) from fetal porcine SDSCs [8]. Subsequent studies demonstrated that SDSCs with germ cell potential could be generated from neonatal mouse skin [7, 15, 16]. Likewise, female and male human fetal SDSCs were differentiated into germ cell-like cells (GCLCs) able to initiate defective meiosis in vitro [17]. Intriguingly, aggregates consisting of OLCs and ovarian somatic cells derived from mouse SDSCs were able to secrete estradiol and progesterone in response to gonadotropin stimulation and to restore endogenous serum estradiol levels when transplanted in ovariectomized females [16]. Such OLCs were generated from CD34-positive hair follicle stem cells (HFSCs) isolated from early postnatal mice [6]. However, the derivation of GCLCs from SDSCs remains an inefficient process restricting its applicative possibilities. In addition, the cellular origin of SDSCs generated from neonatal skin remains elusive.
Using single-cell RNA sequencing (scRNA-seq) we recently analyzed cell fate commitment during in utero skin development in mice and goats [18, 19]. A single-cell transcriptome atlas of neonatal mouse skin is now available providing a reference for understanding cellular heterogeneity of SDSCs cultured in vitro.
In the present study, we use scRNA-seq to dissect the cellular heterogeneity of SDSCs generated in vitro from neonatal skin and unveil their cellular origin from multipotent papillary dermal fibroblast progenitors (pDFPs). Utilizing a modified protocol, adapted from those described to induce functional gametes from mouse pluripotent stem cells, we show that these cells could be efficiently induced to give rise to primordial germ cell-like cells (PGCLCs) in vitro. Furthermore, PGCLCs can be induced into spermatogonia stem cell-like cells (SSCLCs) in vitro and these SSCLCs showed meiotic potential when cultured with testicular organoids.
Materials and methods
Experimental animals
All animals used in the current study were housed and maintained in a temperature (22–24 °C) and light (12 h light and 12 h dark) controlled room with ad libitum access to food and water. CAG/eGFP transgenic mice (Stock TgN (GFPU) 5 Nagy) strain with ubiquitously expressed eGFP (kindly provided by Dr. Xiao Yang from the Institute of Biotechnology, Beijing), were used for monitoring SSCLCs differentiated from the SDSCs-generated PGCLCs in reconstituted testicular organoids. CD-1 mice were used for MEF feeder cell preparation and testicular somatic cell isolation and were purchased from the Vital River Laboratory Animal Technology Co., Ltd, Beijing. Mice were killed by decapitation and about 120 CAG/eGFP transgenic mice and 10 CD-1 mice were involved in the current study. Experimental animal management was approved by the Ethics Committee of Qingdao Agricultural University (Approval Number: 2019-021).
Dissociation of skin and generation of SDSC spheres
The skin dissociation procedure used here is described in detail in [7]. Briefly, the dorsal skin of newborn CAG/eGFP transgene male mice was isolated using surgical scissors following euthanasia. After removing the fat tissue and contaminating blood with forceps, the tissues were cut into 1 mm2 pieces with scissors. Dissociation was then performed with trypsin–EDTA (0.25%) solution (Sorlabio, T1300, Beijing, China) by incubating for 25 min at 37 °C. After dissociation, cells were dispersed by mechanical pipetting, and a single-cell suspension was obtained using a cell strainer (40 μm, Biologix, 15-1040, Grand Island, NY, USA). The obtained single cells were then cultured in DMEM/F12 (Thermo Fisher Scientific, 8121453, Beijing, China) medium containing 40 ng/ml basic fibroblast growth factor (bFGF, PeproTech, 100-18B, Rocky Hill, NJ, USA), 1 × B-27 (Gibco, 17504044, Grand Island, NY, USA), 20 ng/ml epidermal growth factor (EGF, R&D Systems, 2028-EG, Minneapolis, MN, USA), and 1% penicillin/streptomycin (Solarbio, P1400, Beijing, China) in culture dishes for cells growing in suspension (Sarstedt, 83.3902.500, Nümbrecht, Germany) at 37 °C, 5% CO2. Medium was changed every four days and the generated SDSC spheres were harvested after 12 days of culture for further procedures.
Preparation of MEF feeder cells
Mitotically inactivated mouse embryonic fibroblast (MEF) feeder cells were obtained following standard procedures as described in Curr. Protoc. Stem Cell Biol. 3:1C.3.1-1C.3.17. © 2007 by John Wiley & Sons, Inc.
Generation of PGCLCs
A two-step procedure was used to generate PGCLCs from SDSCs. First, in order to produce epiblast cell like cells (EpiLCs), P2 SDSC spheres were mechanically dispersed into a single-cell suspension and seeded at 5 × 105 cells/well into a 24-well plate for cells growing in suspension. The EpiLC induction medium was M199 supplemented with 0.5% insulin-transferrin-selenium (ITS, Thermo Fisher Scientific, 41400045, Waltham, MA, USA), 0.1% bovine fetuin (Merck Millipore, 341506, Palmerston North, New Zealand), 1.5% bovine serum albumin (BSA, Sigma, B2064), 30 ng/ml BMP4 (R&D Systems, 314-BP-050), 0.23 mM sodium pyruvate (Gibco, 11360-070), 1 ng/ml EGF in media medium 199 (Thermo Fisher Scientific, 11150067, Beijing, China). The medium was changed every two days and the first EpiLC colonies were observed after 12 h of culture. In the second step, to induce PGCLCs, colonies of EpiLCs on day 4 were dissociated into single cells by pipetting and plated onto mitotically inactivated MEF feeder cells at a concentration of 5 × 105 cells/ml. The PGCLC differentiation medium was DMEM/high glucose (Gibco, C11995500BT), supplemented with 10% FBS, 40 ng/ml bFGF, 40 ng/ml SCF (R&D Systems, 455-MC-010), 20 ng/ml EGF, 1% MEM non-essential amino acids solution (Thermo Fisher Scientific, 11140-050, Waltham, MA, USA), 1% GlutaMAX™ Supplement (Thermo Fisher Scientific, 35050061, Waltham, MA, USA), 1% sodium pyruvate, 50 μM β-mercaptoethanol (Sigma, M-7522), and 1% penicillin/streptomycin. Half medium was changed every two days being careful not to remove suspended cells.
Differentiation of SSCLCs from PGCLCs
For SSCLCs differentiation from PGCLCs, we used a testicular organoid culture system termed the three-layer gradient system previously described [20] with some modifications. Briefly, testes dissected from 7 days postpartum CD-1 mice were cut into small pieces and a monodispersed cell population was obtained by subsequent incubation in 2 mg/ml collagenase IV solution (Sigma, C5138) for 15 min and TrypLE™ Express enzyme for 10 min at 37 °C, followed by repeated pipetting in DMEM. Cells were then resuspended in SSCLC induction medium and plated into a 10 cm tissue culture dish treated with 0.2% (w/v) gelatin. Not adherent cells (mainly germ cells) were removed through aspirating the supernatant after 4, 12, and 24 h in vitro culture while the somatic cells attached at the bottom of the culture dish were harvested at 24 h and combined with 8–10 day induced eGFP-PGCLCs (cell ratio 1:1) at a concentration of 4 × 107 cells/ml. The cell suspension was then mixed with Matrigel (Corning, 356231) at a ratio of 1:1 and about 16 μl of this mixture was transferred onto a layer of 1.5% agarose gel (Tsingke Biotechnology, TSJ001, Beijing, China) applied to the bottom of a 24-well plate. The plate was transferred to the incubator for 15 min at 37 °C for gelification. Finally, 400 μl of SSC induction medium was added to the well and the culture was carried out in a 5% CO2 humidified incubator at 37 °C. The SSC induction medium was referenced from Ishikura et al. [21] and consisted of StemPro-34 SFM medium (Gibco, 10640-019) supplemented with 20 ng/ml EGF, 1 × insulin-transferrin-selenium, 10 ng/ml bFGF, 1 μl/ml DL-lactic acid (Sigma, 69785), 2 mM L-glutamine, 6 mg/ml d-( +)-glucose (Sigma, G8270), 30 μg/ml pyruvic acid (Sigma, 107360), 5 mg/ml BSA, 60 ng/ml progesterone (Sigma, P0130), 1 × MEM vitamin solution (Gibco, 11120-052), 1 × non-essential amino acids, 30 ng/ml β-estradiol (Sigma, E8875), 50 μM β-mercaptoethanol, 100 nM ascorbic acid (Sigma, A4544), 1% FBS, 10 μg/ml d-biotin (Sigma, B-3399), and 10 ng/ml glial cell line-derived neurotrophic factor (GDNF, R&D Systems, 212-GD-050).
Immunofluorescence and immunohistochemistry
For immunofluorescence (IF), cell clusters or dissociated cells were fixed with 4% paraformaldehyde (Sorlabio, P1110) on ice for 30 min. Then, the cells were transferred to glass slides for drying. Samples were then permeabilized with 0.5% Triton X-100 in PBS (Sorlarbio, T8200) for 10 min, and incubated with blocking buffer (permeabilization containing 10% goat blocking serum, BOSTER, AR0009, Wuhan, China) for 30 min. After blocking, the samples were then incubated with the corresponding primary antibodies overnight at 4 °C. Secondary antibodies were added according to the species source of primary antibodies incubation carried out at 37 °C for 2 h. Vectashield mounting media (Vector Laboratories, H-1000, Burlingame, CA, USA) for mounting and a confocal microscope (Leica, TCS SP5 II, Wetzlar, Germany) for image acquisition were used at 20x. Immunohistochemistry (IHC) of skin sections was performed using the procedures we recently described [18]. All antibodies used are listed in Additional file 1: Table S1.
Bisulfite sequencing
Bisulfite sequencing was performed as previously described [22, 23]. Briefly, the TIANGEN Micro DNA isolation Kit (Tiangen, DP316, Beijing, China) was used to extract genomic DNA from PGCLCs. Bisulfite treatment was then performed using an EpiTect® Bisulfite kit (EpiTect, 59104, Hilden, Germany). Differentially methylated regions of Igf2r and Peg3 genes were then amplified by RT-PCR as previously described [24] and the target sequences cloned into the pDM19-T Vector (TaKaRa, D102A, Dalian, China) for downstream sequencing.
Gene expression analysis
Total RNA was extracted from cell aggregates using SPARKeasy Tissue/Cell RNA Rapid Extraction Kit (SPARKjade, AC0202, Shandong, China) and reverse transcription was performed using a SPARKScript RT plus Kit (SPARKjade, AG0304). A Light Cycler SYBR Green I Master (Roche, 04707516001, Mannheim, Germany) kit was used to prepare PCR mix and PCR was performed using a Light Cycler 480 Real-Time PCR System (Roche, Mannheim, Germany). The relative gene expression was calculated using the 2−(△△Ct) method and Gaphd was used as a housekeeping gene. Primer sequences used are listed in Additional file 2: Table S2.
Western blotting analysis
Cells were firstly harvested by centrifugation and lysed using RIPA lysis solution (Beyotime, P0013B, Beijing, China). The extracts were then analyzed by agarose gel electrophoresis with 12% SDS-PAGE, at 80 V for 20 min, and at 120 V for 2.5 h. Bio-Rad Trans-Blot (Bio-Rad, Hercules, California, USA) was used to transfer the proteins onto the polyvinylidene difluoride membranes (Millipore, ISEQ00010, Bedford, MA, USA). Blocking was performed with PBST containing 5% BSA on ice for 4 h and the membranes were incubated with primary and secondary antibodies for 4 h and overnight, respectively. BeyoECL Star Chemiluminescence Kit (Beyotime, P0018S, Beijing, China) was used for protein band detection. Quantification of protein blots was performed using AlphaView SA software (Alpha Innotech Corporation, San Leandro, CA, USA) according to the user guide using default parameters.
Meiotic chromosome spread
Meiotic chromosome spread staining was performed as we previously described [25]. Briefly, testicular organoid tissues were harvested with a 1 ml pipette and transferred in a hypotonic solution containing 30 mM Tris (Sorlabio, T8060), 50 mM sucrose (Sorlabio, S8271), 17 mM citric acid (Sorlabio, C8610), 5 mM EDTA (Sorlabio, E8040), 2.5 mM dl-dithiothreitol (Sorlabio, D8220) and 1 mM phenylmethanesulfonyl fluoride (Sorlabio, IP0280) in deionized water for 45 min on ice. The samples were then transferred to a 4% paraformaldehyde solution and were dispersed using a pair of precision forceps before spreading evenly on a glass slide. After overnight at room temperature, the slides were washed with a wetting agent (Kodak, 1464502, Rochester, NY, USA) and then treated with the blocking buffer containing 1% goat serum in PBS for 1 h at room temperature. The samples were further incubated with the corresponding primary antibodies at 37 °C overnight. After careful washing, the matched secondary antibodies were added, and incubation was carried out at 37 °C for 2 h. At last, the slides were incubated in Hoechst 33342 (Beyotime, C1017) or DAPI (Beyotime, C1002) solution to stain nuclei for 5 min at room temperature.
Single-cell cDNA library preparation and sequencing
We used 10 × Genomics’ Chromium Single Cell 3' V3 Gel Beads Kit (10 × Genomics, PN-1000075, Pleasanton, CA, USA) to construct a single-cell cDNA library, and all procedures were performed according to the recommended protocol. Briefly, cells with cell viability higher than 90% were projected for cell capture using a 10 × Genomics Chromium controller (10 × Genomics, Pleasanton, CA). Sequencing was performed by Novogene, Inc.
10 × Genomics scRNA-seq data preprocessing
Alignment and quantification of UMI counts were performed using the CellRanger (v3.0.2) software according to the standard protocol (https://www.10xgenomics.com/). After that, the processed expression matrix was then analyzed using the Seurat (v3) R package for downstream quality control and visualization [26]. For quality control, cells were filtered out using the default parameter in Seurat and data integration was performed with FindIntegrationAnchors function in the Seurat package. Dimension reduction and cluster identification were performed with RunUMAP and FindClusters functions with the following parameters: dims = 1:25, resolution = 0.8. Other parameters were set as default according to the official instructions.
RNA velocity analysis and single-cell trajectory
To determine the directionality of differentiation, the velocyto package was used to infer velocity vectors in single cells according to the published protocol [27]. Briefly, the spliced and unspliced count metrics were firstly generated directly from the Cellranger output file using velocyto run10x function using the standard procedure. The obtained metric files were further analyzed using scVelo package [28], and the velocities were projected onto pre-computed UMAP embedding using scv.pl.velocity_embedding_stream function with default parameters.
For single-cell trajectory inference, the slingshot R package was used to infer dynamic gene expression along pseudotime [29]. The root cell cluster was determined according to the directionality of RNA velocity vectors incorporated in the UMAP plot as previously described [30].
Gene ontology (GO) enrichment
We used Metascape to perform GO enrichment analysis and the analysis procedure was performed according to the online tutorial (https://metascape.org/) [31].
Statistical analysis
Statistical tests were analyzed using the Graphpad Prism 7 software. Statistical significance was analyzed with the Student’s unpaired t-test or one-way ANOVA test.
Results
Dissection of cellular heterogeneity of P2 SDSCs and newborn mouse skin at single-cell resolution
All SDSCs described in the current study were isolated from newborn male pup skin according to published protocols [7, 8, 32]. Transgenic mice carrying a ubiquitously expressed enhanced green fluorescent protein (CAG/eGFP) were used to generate SDSCs in order to obtain traceable cells for the experiments involving the culture of SSCLCs within testicular organoids (Additional file 3: Fig. S1A).
scRNA-seq was used to analyze the cellular heterogeneity in newborn skin and SDSCs cultured to passage two (P2). We generated 8000 single-cell transcriptome profiles in total (Additional file 3: Fig. S1B), and after rigorous quality control (Additional file 3: Fig. S1C), 7543 single-cell transcriptome profiles were subjected to transformations into low-dimensional spaces by uniform manifold approximation and projection (UMAP). After batch correlation, we identified a total of 17 cell clusters of which 7 were mainly comprised of cell populations derived from SDSCs (Fig. 1A, B). Hierarchical clustering indicated that 6 of these, namely 0, 1, 2, 5, 6, and 7, clustered in the same branch consistent with their origin from SDSCs, while cluster 9 clustered with the other skin-derived cell types (Fig. 1C).
Analysis of single-cell transcriptome profiles of newborn mouse skin and P2 SDSCs. A UMAP projection of 7543 single-cell profiles reveals 17 cell clusters in newborn skin and SDSCs. B UMAP plot color-coded by cell source. C Hierarchical clustering of different cell clusters identified by UMAP analysis. D Representative canonical cell type-specific marker expression across all cell clusters. E Percentage of different cell clusters in newborn skin and SDSCs
To further define the cell identities in the UMAP plot we annotated the different cell clusters using a series of canonical marker genes [18]. On the basis of the higher expressed genes, nine different cell types were identified as follows: SDSC clusters 0, 1, 2, 5, 6, 7 and 9 showing high expression of the dermal cell lineage gene markers Ptn, Col1a1 and Lum were designated as dermal fibroblast (DF) populations, cluster 8 (Krt5 and Perp) as hair shaft cells, cluster 10 (Pecam1 and Kdr) as endothelial cells, cluster 11 (Wnt7b and Krt16) as inner root sheath cells (IRS), cluster 12 (Dct and Pax3) as muscle cells, cluster 13 (Cd52 and Fcer1g) as immune cells, cluster 14 (Plp1 and Fabp7) as melanocytes, cluster 15 (Rgs5 and Myh11) as pericytes and cluster 16 expressing Prdm1 and Sox18, as dermal papillae cells (DP) (Fig. 1D and Additional file 3: Fig. S1D). The comparison of the cluster composition between newborn mouse skin tissues and P2 SDSCs indicated that SDSCs cells were heterogeneous, and mainly consisted of dermal lineage-derived cells (Fig. 1E).
Reconstruction of cellular differentiation trajectory and RNA velocity analysis unveil the origin of SDSCs
Since it is known that in the neonatal skin reside dermal fibroblasts (DFs) with distinct differentiation potential, including papillary, reticular and hypodermal fibroblasts [33], we next investigated DF lineages ongoing in the in vitro cultured SDSCs. In order to capture the differentiation trajectories of DFs, we utilized RNA velocity analysis that is able to predict cell fate on the basis of the ratio in scRNA data between spliced and un-spliced mRNA and is considered a robust tool to identify cell fate commitment in heterogeneous single cell RNA-seq data [27]. Noteworthy, we found that the projection of RNA velocity vectors onto UMAP plots showed two distinct differentiation directories (Fig. 2A). Clusters 0 and 6 were root states, hence representing cells at the beginning of differentiation [30], while clusters 2 and 16 represented final differentiation states. From the root states, we reconstructed two pseudotime differentiation trajectories towards the end states to identify gene expression dynamics of cell fates 1 (cluster 2) and 2 (cluster 16). The results showed that cells at final fate state 1 (cluster 1, 2, 6, 7) expressed high levels of Cenpa and Cdc20, genes involved in mitotic cell cycle processes, while the dermal papillae marker genes Lef1 and Ppm1e were expressed at higher levels by cells at the final fate of state 2 (cluster 0, 5, 16) [34] (Fig. 2B, C). We further identified RNA velocity driver genes for both differentiation trajectories (Fig. 2D). For cell fate 1, the top enriched RNA velocity driving genes, ranked by their roles in driving the velocity trajectories [34], included Knstrn which encodes a protein involved in chromosome segregation during mitosis [35], and Miip which encodes a negative regulator of cell migration and mitosis [36]. Moreover, gene ontology (GO) enrichment analysis of the top 100 RNA velocity driver genes showed that “mitotic cell cycle process”, “cell cycle”, and “regulation of mitotic cell cycle”, were the top represented GO terms for cell fate 1 (Fig. 2E). In line with this, heatmap of cluster-specific represented GO terms and a comparison of shared genes and GO identified the expression of cell cycle mitotic genes along this pathway (Additional file 3: Fig. S2A, B). Interestingly, for cell fate 2 (cluster 16) top-ranked RNA velocity genes resulted the same Lef1, the dermal papillae cell fate RNA velocity driving gene identified previously in vivo [34], and Ppm1e, encoding a regulator of AMPK phosphorylation [37]. On this basis, we designated fate 1 as a cell renewal state and fate 2 as the differentiation into the papillary fibroblast lineage. Collectively, these data demonstrated that SDSCs mainly consist of papillary dermal fibroblast progenitors (pDFPs) able to self-renewal and differentiation into papillary DFs in vitro.
Combined RNA velocity and trajectory inference unveil the cellular origin of SDSCs. A Projection of RNA velocity vectors in the UMAP plot. B Slingshot infers the pseudotime trajectory in the P2 SDSCs. C Expression of cell fate 1 and cell fate 2 representative marker genes along pseudotime trajectories. D Expression of top 5 cell cluster-specific RNA velocity genes; genes were ranked by their roles in driving the velocity trajectories. E GO enrichment analysis of top 100 RNA velocity genes in the end state of cell fate 1 and 2. F Expression of top 5 cell fate 2 RNA velocity genes in the skin of E14.5 foetuses and 5 dpp newborn skin
To further validate our RNA velocity analysis, we retrieved mRNA expression data from Rezza et al. [38] and assessed the expression of the top 5 RNA velocity genes in the endpoint of cell fate 2 (Lefty, Ppm1e, Spata6, Glis1, Dlk1) in the skin of E14.5 embryos and 5 dpp pups (Fig. 2F) [39]. Such analysis established that these genes were mainly expressed in the E14.5 skin dermal cells (DCs) and showed higher expression in the DF and DP cells in the 5 dpp pup skin. This validated our RNA velocity analysis and confirmed that SDSCs (hereafter referred to as pDFPs) mainly comprise papillary dermal fibroblast progenitors able to differentiate into DP cells [33].
EGF promotes pDFP proliferation in vitro
Since the enriched GO terms of cell fate 2 RNA velocity driving genes analysis also included the “EGFR1 signaling pathway”, we then tested if EGFR signaling was active and has a role in the cultured pDFPs. We first confirmed by immunohistochemistry that EGFR was expressed in the skin of E16.5 fetuses and 0 dpp pups in epidermal cells, hair follicles, and dermal fibroblasts (Fig. 3A). Next removing EGF from the culture medium, we found that the ability of pDFPs to generate large colonies was significantly reduced (Fig. 3B). Collectively, these results further confirmed that SDSCs consist of multipotent pDFPs, and EGF signaling is pivotal for pDFPs colony formation in vitro.
EGF promotes proliferation and formation of large pDFPs spheres (colonies) in vitro. A IF staining of EGFR in epidermal cells, hair follicles and dermal fibroblasts of the skin of E16.5 fetuses and 0 dpp pups; dotted lines indicate dermal and epidermal interface, and black arrows indicate papillary dermis. Pictures were taken using an optical microscope (20x, Olympus BX51, Japan). Scale bars, 100 μm. B Effects of EGF on the diameter of P2 pDFP colonies. Pictures were taken using an eclipse microscope (10x, Nikon TE2000, Japan)
Specification of PGCLCs from pDFPs in vitro
We next investigated the germline potential of pDFPs and designed an optimized culture strategy to generate primordial germ cell-like cells (PGCLCs) from these cells. In order to efficiently generate PGCLCs from pDFPs we established a different protocol from those previously published using SDSCs [7, 8, 32]. In general, differentiation of PGCLCs from stem cells involves two main steps, induction of epiblast-like cells (EpiLCs) followed by PGCLC specification (Fig. 4A). On this basis in the present paper, eGFP-pDFPs were dissociated into single-cell pellets and were subjected to EpiLC induction under floating conditions in the presence of various concentrations of bone morphogenetic protein 4 (BMP4) (Additional file 3: Fig. S3A). The results showed that 30 ng/ml BMP4 promoted the formation of the largest eGFP-colonies both on day 1 and day 3 of culture (Additional file 3: Fig. S3B). These colonies can be considered bona fide EpiLCs since after real-time RT-PCR analyses they were shown to express a high level of the epiblast marker genes Dnmt3a, Dnmt3b and Wnt3a [7] (Additional file 3: Fig. S3C).
PGCLC differentiation from pDFPs. A Scheme showing PGCLC and SSCLC induction from SDSCs. B Representative morphology of PGCLCs generated from pDFPs at increasing culture times. Pictures were taken using an eclipse microscope (10x, Nikon TE2000, Japan). Scale bars, 100 μm. C Expression of the germ cell marker DDX4 and DAZL in day 6 eGFP positive pDFP-derived PGCLCs. Pictures were taken using an confocal microscope (20x, Leica, TCS SP5 II, Germany). Scale bars, 25 μm. D Expression of genes proper of specified PGCs by day 2, 4, 6 pDFP-derived PGCLCs. E Western blotting analysis of H3K27me3 and H3K9me2 expression during PGCLCs differentiation from pDFPs. F Bisulfite sequencing of DMRs of the imprinted genes Igf2r and Peg3 in P2 SDSCs, day 6 pDFP-derived PGCLCs and E12.5 PGCs
eGFP labelled EpiLCs were next dissociated into single cells and seeded onto MEF cells for PGCLC specification in the induction medium containing 30 ng/ml bone morphogenetic protein 4 (BMP4), 40 ng/ml stem cell factor (SCF), 40 ng/ml bFGF and 20 ng/ml EGF (Fig. 4B). After 4 days of differentiation, eGFP-positive round shiny cells could be observed on the surface of the feeder cells (Additional file 3: Fig. S3D). In subsequent days, these cells proliferated rapidly and gradually gave rise to floating clusters that stained positively for the two canonical late PGC markers DDX4 (also known as VASA) and DAZL [40] (Fig. 4C). The identity of these cells as a mixed population of early and late PGCLCs was further confirmed by the increased expression of the germ cell marker genes including Ddx4, Dazl, Blimp1 (also known as Prdm1), Dppa3 (also known as Stella), and Prdm14 during prolonged in vitro culture (Fig. 4D).
The proper histone modifications and epigenetic reprogramming of PGCs are crucial for germ cell lineage specification [41], we then investigated whether histone modifications and epigenetic reprogramming proceeded properly in PGCLCs derived from pDFPs. Western blotting results showed that both H3 lysine 27 trimethylation (H3K27me3) and histone H3 lysine 9 dimethylation (H3K9me2) undergo progressive erasure and increase, respectively, from pDFPs to 10d PGCLCs showing a tendency to increase (H3K27me3) and erasure (H3K9me2) in 20d PGCLCs resembling the changes occurring in specified PGCs in vivo (Fig. 4E). The bisulfite sequencing of the differentially methylated regions (DMRs) of the imprinted Igf2r and Peg3 loci showed lower levels of DNA methylation in P14 PGCLCs compared with P2 pDFPs, consistent with DNA methylation erasure typical of late PGCs in vivo. This process, however, appeared less efficient in PGCLCs than in their in vivo counterparts (Fig. 4F).
Generation of SSCLCs from PGCLCs within testicular organoids
We then tested whether it was possible to induce pDFP-derived PGCLCs into spermatogonial stem cell-like cells (SSCLCs) as recently described from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs)-derived PGCLCs [21]. To this aim, we first changed the PGC induction medium with the SSC induction medium formulated by Ishikura et al., and mixed the eGFP-PGCLCs with somatic cells obtained from prepubertal testis to generate testicular organoids as described [21] with some modifications (see details in the Materials and Methods section).
Beginning at day 4 of culture, cordonal structures resembling seminiferous cords containing eGFP-positive cells were observed (Fig. 5A). After dissociation of these structures at 14 days of culture, immunofluorescence staining for the canonical spermatogonia stem cell (SSC) proteins PLZF transcription repressor and GDNF receptor GFRa1 [42, 43], showed a significant number of double-positive eGFP-positive cells (Fig. 5B), thus suggesting their SSCLC identity. Finally, we investigated if these cells were able to enter meiosis within the cordonal structures. To this aim, the testicular organoid tissues were dissociated after 2, 4, 5, and 8 days of culture (Additional file 3: Fig. S4), and chromosome spreads were then performed to investigate whether these SSCLCs can initiate meiotic programs under in vitro conditions. Noteworthy, entering into meiosis of pDPFs-derived SSCLCs was further supported by chromosome spreads of testicular organoid tissues cultured for 8 days showing eGFP cells IF positive, in addition to SYCP3, and the two meiotic prophase proteins γH2AX and RAD51 (Fig. 5C) [31]. However, eGFP positive cells stained for the meiotic synapse protein SYCP1 and the recombination protein MLH1 were not found.
Induction of SSCLCs from pDFP-derived PGCLCs. A Representative pictures of day 7 and 14 aggregates of eGFP-PGCLCs and somatic testicular cells; at 14 days, testicular organoids showing structures resembling testis cords containing eGFP-cells are recognizable. Pictures were taken using an eclipse microscope (10x, Nikon TE2000, Japan). Scale bars, 50 μm. B Expression of the canonical SSC marker PLZF and GFRα1 in eGFP-cells isolated from testicular organoids after 14 days of culture. Pictures were taken using a confocal microscope (20x, Leica, TCS SP5 II, Germany). Scale bars, 20 μm. C Meiotic chromosome spreads of testicular tissues from 8 dpp testes of CAG/eGFP mice and testicular organoids containing eGFP-SSCLCs after 8 days of culture stained with antibodies against meiotic proteins; note that positive staining for SYCP3, γH2AX, and RAD51 was observed in all samples while positive cells for SYCP1 and MLH1 were observed only in the control samples from 8 dpp testes of CAG/eGFP mice. Pictures were taken using a confocal microscope (20x, Leica, TCS SP5 II, Germany). Scale bars, 20 μm
Discussion
The main results achieved in the present paper were the identification of the cellular origin of the SDSCs from the skin of newborn mice and their efficient differentiation into cells of the male germ cell line.
Since in vitro isolation of multipotent stem cells from skin in early 2000 [11,12,13,14], the cells able to give rise to SDSCs have remained elusive. These cells were reported to reside in the dermis and, in principle, could be members of one or more of the stem cell populations residing in the hair follicles (bulge, dermal papillae and dermal sheath) and skin dermal layer, this latter including papillary, reticular and hypodermal fibroblast progenitors [33]. The lack of conserved lineage markers of these stem cells and the dynamics of the differentiation processes in which they are involved complicated discovering the origin of SDSCs. In the present paper, this question was solved using scRNA-seq and related bioinformatics analyses. It was revealed that SDSCs mainly consist of pDFPs, one of the three fibroblast precursors originating from multipotent dermal fibroblasts in the skin [33].
As reported in the introduction, in vitro SDSCs can be induced with distinct protocols to give rise to neuroectodermal and mesodermal progenies, including glia, adipocytes, neurons, smooth muscle cells, and, surprisingly, to cells of the germ cell lineage such as PGCLCs and OLCs [5]. In this regard, we have reported that it was possible to generate OLCs from HFSCs, one of the putative sources of SDSCs residing in the bulge region of the hair follicle and of epithelial derivation (see above), obtained from vibrissa hair follicles of 7 dpp mice [6]. We were, therefore, quite surprised to find that the scRNA-seq indicated pDFPs as the main members of SDSCs generated from the skin of mice of the same age. This suggests heterogenicity of SDSC spheres possibly with different cellular origins and prevalence of pDFPs when whole skin is used for their generation.
If SDSCs of distinct cellular origin also possess different GCLC potential remains to be determined. So far, however, the efficiency of the SDSC differentiation process into PGCLCs and OLCs and the quality of the GCLCs generated have been quite modest. As matter of fact, only mouse ESCs or iPSCs have been reported to generate germ cells able to complete spermatogenesis and oogenesis and give rise to functional gametes in vitro [3, 4]. The pluripotency of these stem cells and the induction to the EpiLC phenotype, seem two crucial conditions to generate male or female PGCLCs that are inducible into functional gametes. In any case, the production of functional sperm and oocytes in vitro requires inserting PGCCLs within reconstituted testes or ovaries [3, 44, 45]. Although SDSC-derived pDFPs do not possess pluripotency, supposing that they are probably less ‘biased’ than other ADSCs, we decided to investigate if subjected to a protocol of transient early mesodermal phenotype induction as that used for ESCs and iPSCs they were able to generate PGCLCs competent to differentiate into SSCs which has not previously been reported. For this last step, we utilized reconstituted testes to recapitulate SSC niche using a recently described three-dimensional multilayer model [20]. The results were promising since SDSC-derived pDFPs following induction into bona fide EpiLCs were efficiently specified into PGCLCs, following by cultured under SSC conditions, these PGCLCs generated SSCLCs which inserted within the reconstituted testicular organoids and appeared to be able to initiate meiosis. However, the generated SSCLCs showed a limited capacity of meiosis initiation. A previous study by Ishikura et al. [21], reported that abnormal epigenetic reprogramming is a limiting factor controlling the successful generation of spermatozoa from pluripotent stem cell-derived GCLCs. In this regard, the bisulfite sequencing analysis indicated that PGCLCs generated from SDSC-derived pDFPs possessed higher methylation levels at the DMRs of the imprinted Igf2r and Peg3 loci compared with their in vivo PGC counterparts, thus suggesting that proper DNA demethylation of specific loci is a key regulatory factor controlling meiotic initiation and progression. This supports the notion that the epigenome of GCLCs derived from ASCs is a key indicator of their capability to correctly enter into meiosis. How establishing correct epigenetic reprogramming in ASC-derived GCLCs remains a crucial task for future studies. Key challenges ahead include of course reconstituting the entire spermatogenesis in vitro and in vivo from these GCLCs, but promising results have been already obtained using mouse ESCs [4].
Conclusions
Here, by utilizing scRNA seq, we demonstrated that SDSCs derived from multipotent papillary dermal fibroblast progenitors (pDFPs) residing in the dermal layer show germline potential in vitro, and EGF signaling is pivotal for the capability of these progenitors to proliferate and form large colonies in vitro. More importantly, we established an optimized protocol to efficiently generate primordial germ cell-like cells (PGCLCs) from SDSCs. Together, these findings here lay the foundation for future generating functional gametes from adult stem cells.
Availability of data and materials
scRNA seq data of SDSCs are retrieved from our previous published dataset and the data are deposited in the NCBI GEO databases (https://www.ncbi.nlm.nih.gov/geo/) under Accession Number: GSE131498. scRNA seq data of SDSCs are deposited in the GSA database (https://bigd.big.ac.cn/) under Accession Number: CRA005534.
Change history
23 July 2024
A Correction to this paper has been published: https://doi.org/10.1186/s13287-024-03838-6
Abbreviations
- SDSCs:
-
Skin-derived stem cells
- scRNA-seq:
-
Single-cell RNA sequencing
- pDFPs:
-
Papillary dermal fibroblast progenitors
- EGF:
-
Epidermal growth factor
- PGCLCs:
-
Primordial germ cell-like cells
- SSCLCs:
-
Spermatogonial stem cell like cells
- EpiLCs:
-
Epiblast cell like cells
- ESCs:
-
Embryonic stem cells
- iPSCs:
-
Induced pluripotent stem cells
- ASCs:
-
Adult stem cells
- GCLCs:
-
Germ cell-like cells
- EPSCs:
-
Epidermal stem cells
- M-S-Cs:
-
Mesenchymal stem cells
- OLCs:
-
Oocyte-like cells
- HFSCs:
-
Hair follicle stem cells
- MEF:
-
Mouse embryonic fibroblast
- IF:
-
Immunofluorescence
- GO:
-
Gene ontology
- UMAP:
-
Uniform manifold approximation and projection
- DFs:
-
Dermal fibroblast
- DMR:
-
Differentially methylated regions
- SSC:
-
Spermatogonia stem cell
- RA:
-
Retinoic acid
References
Mouka A, Tachdjian G, Dupont J, Drevillon L, Tosca L. In vitro gamete differentiation from pluripotent stem cells as a promising therapy for infertility. Stem Cells Dev. 2016;25(7):509–21.
Ge W, Chen C, De Felici M, Shen W. In vitro differentiation of germ cells from stem cells: a comparison between primordial germ cells and in vitro derived primordial germ cell-like cells. Cell Death Dis. 2015;6(10):e1906.
Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, et al. Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature. 2016;539(7628):299–303.
Ishikura Y, Ohta H, Sato T, Murase Y, Yabuta Y, Kojima Y, et al. In vitro reconstitution of the whole male germ-cell development from mouse pluripotent stem cells. Cell Stem Cell. 2021;28:2167–79.
Ge W, Cheng SF, Dyce PW, De Felici M, Shen W. Skin-derived stem cells as a source of primordial germ cell- and oocyte-like cells. Cell Death Dis. 2016;7(11):e2471.
Sun YC, Ge W, Lai FN, Zhang RQ, Wang JJ, Cheng SF, et al. Oocyte-like cells induced from CD34-positive mouse hair follicle stem cells in vitro. J Genet Genom. 2017;44(8):405–7.
Sun R, Sun YC, Ge W, Tan H, Cheng SF, Yin S, et al. The crucial role of Activin A on the formation of primordial germ cell-like cells from skin-derived stem cells in vitro. Cell Cycle. 2015;14(19):3016–29.
Dyce PW, Wen L, Li J. In vitro germline potential of stem cells derived from fetal porcine skin. Nat Cell Biol. 2006;8(4):384–90.
Linher K, Dyce P, Li J. Primordial germ cell-like cells differentiated in vitro from skin-derived stem cells. PLoS ONE. 2009;4(12):e8263.
Danner S, Kajahn J, Geismann C, Klink E, Kruse C. Derivation of oocyte-like cells from a clonal pancreatic stem cell line. Mol Hum Reprod. 2007;13(1):11–20.
Toma JG, Akhavan M, Fernandes KJ, Barnabe-Heider F, Sadikot A, Kaplan DR, et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat Cell Biol. 2001;3(9):778–84.
Dyce PW, Zhu H, Craig J, Li J. Stem cells with multilineage potential derived from porcine skin. Biochem Biophys Res Commun. 2004;316(3):651–8.
Jinno H, Morozova O, Jones KL, Biernaskie JA, Paris M, Hosokawa R, et al. Convergent genesis of an adult neural crest-like dermal stem cell from distinct developmental origins. Stem Cells. 2010;28(11):2027–40.
Rodrigues RM, De Kock J, Branson S, Vinken M, Meganathan K, Chaudhari U, et al. Human skin-derived stem cells as a novel cell source for in vitro hepatotoxicity screening of pharmaceuticals. Stem Cells Dev. 2014;23(1):44–55.
Dyce PW, Liu J, Tayade C, Kidder GM, Betts DH, Li J. In vitro and in vivo germ line potential of stem cells derived from newborn mouse skin. PLoS ONE. 2011;6(5):e20339.
Park BW, Pan B, Toms D, Huynh E, Byun JH, Lee YM, et al. Ovarian-cell-like cells from skin stem cells restored estradiol production and estrus cycling in ovariectomized mice. Stem Cells Dev. 2014;23(14):1647–58.
Ge W, Ma HG, Cheng SF, Sun YC, Sun LL, Sun XF, et al. Differentiation of early germ cells from human skin-derived stem cells without exogenous gene integration. Sci Rep. 2015;5:13822.
Ge W, Tan SJ, Wang SH, Li L, Sun XF, Shen W, et al. Single-cell transcriptome profiling reveals dermal and epithelial cell fate decisions during embryonic hair follicle development. Theranostics. 2020;10(17):7581–98.
Ge W, Zhang W, Zhang Y, Zheng Y, Li F, Wang S, et al. A single-cell transcriptome atlas of cashmere goat hair follicle morphogenesis. Genom Proteom Bioinform. 2021;19:437–51.
Alves-Lopes JP, Soder O, Stukenborg JB. Testicular organoid generation by a novel in vitro three-layer gradient system. Biomaterials. 2017;130:76–89.
Ishikura Y, Yabuta Y, Ohta H, Hayashi K, Nakamura T, Okamoto I, et al. In vitro derivation and propagation of spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Rep. 2016;17(10):2789–804.
Fu XF, Yang F, Cheng SF, Feng YN, Li L, Dyce PW, et al. The epigenetic modifications and the anterior to posterior characterization of meiotic entry during mouse oogenesis. Histochem Cell Biol. 2017;148(1):61–72.
Chao HH, Zhang XF, Chen B, Pan B, Zhang LJ, Li L, et al. Bisphenol A exposure modifies methylation of imprinted genes in mouse oocytes via the estrogen receptor signaling pathway. Histochem Cell Biol. 2012;137(2):249–59.
Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79(4):530–8.
Ge W, Wang JJ, Zhang RQ, Tan SJ, Zhang FL, Liu WX, et al. Dissecting the initiation of female meiosis in the mouse at single-cell resolution. Cell Mol Life Sci. 2021;78(2):695–713.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902.
La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–8.
Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408–14.
Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19(1):477.
Frede J, Anand P, Sotudeh N, Pinto RA, Nair MS, Stuart H, et al. Dynamic transcriptional reprogramming leads to immunotherapeutic vulnerabilities in myeloma. Nat Cell Biol. 2021;23(11):1199–211.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
Shen W, Park BW, Toms D, Li J. Midkine promotes proliferation of primordial germ cells by inhibiting the expression of the deleted in azoospermia-like gene. Endocrinology. 2012;153(7):3482–92.
Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504(7479):277–81.
Phan QM, Fine GM, Salz L, Herrera GG, Wildman B, Driskell IM, et al. Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. eLife. 2020;9:e60066.
Lee CS, Bhaduri A, Mah A, Johnson WL, Ungewickell A, Aros CJ, et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat Genet. 2014;46(10):1060–2.
Wang Y, Wen J, Zhang W. MIIP, a cytoskeleton regulator that blocks cell migration and invasion, delays mitosis, and suppresses tumorogenesis. Curr Protein Pept Sci. 2011;12(1):68–73.
Voss M, Paterson J, Kelsall IR, Martin-Granados C, Hastie CJ, Peggie MW, et al. Ppm1E is an in cellulo AMP-activated protein kinase phosphatase. Cell Signal. 2011;23(1):114–24.
Rezza A, Wang Z, Sennett R, Qiao W, Wang D, Heitman N, et al. Signaling networks among stem cell precursors, transit-amplifying progenitors, and their niche in developing hair follicles. Cell Rep. 2016;14(12):3001–18.
Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, Sicchio C, et al. An integrated transcriptome atlas of embryonic hair follicle progenitors, their niche, and the developing skin. Dev Cell. 2015;34(5):577–91.
Nicholls PK, Schorle H, Naqvi S, Hu YC, Fan Y, Carmell MA, et al. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc Natl Acad Sci USA. 2019;116(51):25677–87.
De Felici M. Nuclear reprogramming in mouse primordial germ cells: epigenetic contribution. Stem cells international. 2011;2011:425863.
Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36(6):647–52.
Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36(6):653–9.
Ishikura Y, Ohta H, Sato T, Murase Y, Yabuta Y, Kojima Y, et al. In vitro reconstitution of the whole male germ-cell development from mouse pluripotent stem cells. Cell Stem Cell. 2021;28(12):2167–79.
Yoshino T, Suzuki T, Nagamatsu G, Yabukami H, Ikegaya M, Kishima M, et al. Generation of ovarian follicles from mouse pluripotent stem cells. Science. 2021;373(6552):eabe0237.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Nature Science Foundation of China (32100683 and 32072734), the Natural Science Foundation of Shandong Province, China (ZR2021QC003), the National Key Research and Development Program of China (2018YFC1003400), the Taishan Scholar Construction Foundation of Shandong Province of China (ts20190946), the Start-up Fund for High-level Talents of Qingdao Agricultural University (6651121003), and the funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
WG and WS conceived the project and designed the experiments. WG, Y-CS, TQ, and C-LC conducted most of the experiment and performed scRNA-seq data analysis. WG, Y-CS, QT, H-XL, T-RH, and C-LC carried out some of the experiments. J-JW, S-FC, P-WD, and M-DF helped with the statistical analyses and provided the technical support. WG and WS wrote the manuscript. P-WD and M-DF edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experimental procedures and animal management procedures were approved by the Ethics Committee of Qingdao Agricultural University (Approval Number: 2019-021; Date of approval: May 16, 2019; Title of the approved project: “Differentiation of germ-cell like cells from skin-derived stem cells”).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Antibodies used in the present study.
Additional file 2: Table S2.
Primers used in the present study.
Additional file 3: Fig. S1.
In vitro propagation of SDSCs and quality control of scRNA seq datasets. A Morphology of eGFP-SDSC spheres formation under the bright and fluorescent field. Pictures were taken using an eclipse microscope (10x, Nikon TE2000, Japan). Scale bars, 50 μm. B Quality matrices of two scRNA-seq datasets revealed by CellRanger. C Comparison of the number of genes detected in each cell (nFeature_RNA), the total number of molecules detected within a cell (nCount_RNA), and the percentage of mitochondria RNA in different datasets. D Representative marker expression in the UMAP plot. Fig. S2. Gene function enrichment comparison of different SDSC clusters. A Heatmap comparing the top enriched GO terms in different cell clusters. B Circos plot representations showing the overlapped GO terms and overlapped genes between different cell clusters. Fig. S3. EpiLC and PGCLC induction. A Effects of different concentrations of BMP4 on the formation of EpiLC colonies. Pictures were taken using an eclipse microscope (10x, Nikon TE2000, Japan). Scale bars, 100 μm. B Statistical comparison of the diameter of EpiLC colonies exposed to different concentrations of BMP4 on day 1 and day 3 of culture. C Expression of the epiblast Dnmt3a, Dnmt3b, and Wnt3a genes during EpiLCs induction revealed by RT-PCR. D Representative bright-field and fluorescent field image of day 4 eGFP-PGCLCs. Pictures were taken using an eclipse microscope (10x, Nikon TE2000, Japan). Scale bars, 100 μm. Fig. S4. Scheme of the testicular organoid setup.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ge, W., Sun, YC., Qiao, T. et al. Murine skin-derived multipotent papillary dermal fibroblast progenitors show germline potential in vitro. Stem Cell Res Ther 14, 17 (2023). https://doi.org/10.1186/s13287-023-03243-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-023-03243-5