Abstract
Introduction
The basis of orthodontic tooth movement (OTM) is the reconstruction of periodontal tissue under stress. Increasing the speed of OTM has always been the focus of attention.
Objectives
Periodontal ligament stem cells (PDLSCs) are direct effector cells of mechanical force, but the mechanism by which PDLSCs sense mechanical stimuli is unclear.
Methods
Human PDLSCs (hPDLSCs) were analyzed in the presence or absence of force loading with the Flexcell loading system in vitro. Then, periodontal tissues were analyzed after mechanical stimulation in vivo. In addition, cells in a confined microenvironment were analyzed to observe changes in the cytoskeleton and migration. Finally, TRPC6−/− mice were used to further verify the effect of TRPC6. After force application, the OTM distance, bone marrow density (BMD), TRPC6 and COL1 expression, and TRAP staining were evaluated in periodontal tissues.
Results
RNA sequencing (RNA-seq) and western blot analyses revealed that TRPC6 was important during mechanical force application to hPDLSCs. Appropriate mechanical force application also induced TRPC6 activation in the OTM model and the confined microenvironment. Under a slightly confined microenvironment, treatment with the TRPC6 inhibitor SKF96365 and TRPC6 knockout decreased the migration speed of hPDLSCs and mouse bone marrow mesenchymal stem cells (mBMSCs). In addition, TRPC6−/− mice showed lower OTM distances and reduced osteogenic and osteoclastic differentiation.
Conclusion
In summary, TRPC6 activation in PDLSCs mediated by appropriate mechanical force application contributes to periodontal tissue reconstruction.
Graphical abstract
PDLSCs modulate periodontal tissue remodeling under appropriate mechanical stimulation through TRPC6; however, under excessive stress, alveolar bone and tooth roots are readily absorbed. Under this condition, environmental factors play a leading role, and the regulatory effect of TRPC6 is not obvious.

Similar content being viewed by others
Introduction
With the gradual increase in people’s awareness of oral health care and aesthetics, orthodontic treatments aiming at health, beauty and orthodontic stability have become a common choice. However, perfect orthodontic treatment is often a long and complex process that usually takes more than two years. Therefore, identifying approaches to improve the speed of OTM has always been the focus of attention. To accelerate OTM, it is important to research the mechanism of periodontal tissue reconstruction under mechanical force.
Mechanical force plays an important role in tissue development and reconstruction and also in OTM [1,2,3,4]. Excessive mechanical force leads to the absorption of alveolar bone and roots, and insufficient force cannot generate the movement of teeth. Therefore, the application of appropriate orthodontic force is very important. The basis of OTM is the reconstruction of periodontal tissue under stress.
In the process of OTM, PDLSCs are the direct effector cells of mechanical force [5]. They sense mechanical stimulation and then translate these signals into biochemical signals to complete the process of periodontal tissue reconstruction [6, 7]. However, the mechanism by which PDLSCs sense mechanical stimulation remains unclear and needs to be further studied.
Mechanosensitive ion channels (MICs) are the main mechanoreceptors on the surface of living cells when a mechanical stimulus is transduced into an intracellular electrical or chemical signal. MICs form a mechanoreceptor system together with the cytoskeleton [8]. MICs mainly include epithelial sodium channels (ENaCs), two-pore potassium channels (K2Ps), transient receptor potential channels (TRPs) and piezo electric ion channels (Piezos).
Transient receptor potential (TRP) calcium channels are mechanosensitive channels and can sense different stimuli [9, 10]. Transient receptor potential channel 6 (TRPC6) is a nonselective cation channel that plays an important role in regulating calcium influx in most nonexcitatory cells [11]. Previous studies have shown that TRPC6 is widely expressed in the brain, kidney, smooth muscle tissue, immune cells and blood cells and is mechanosensitive [12,13,14]. Our previous study showed that TRPC6 played a significant role in the osteogenic differentiation of PDLSCs [15], but the role of TRPC6 in PDLSCs under stress was unclear. Therefore, we hypothesized that PDLSCs are affected by mechanical stimulation, which induces the activation of TRPC6 and further promotes periodontal tissue reconstruction.
Here, using a tensile and compressive force stimulus in vitro and an OTM model in vivo, we assessed the regulatory effect of TRPC6 under stress. In addition, the cells in a confined microenvironment were used to further verify the effect of TRPC6. TRPC6 knockout mice have been used in the pulmonary vasculature [16], so we also used TRPC6−/− mice to further verify the effect of TRPC6 during bone remodeling after appropriate mechanical force application. In conclusion, we show that the activation of TRPC6 in PDLSCs under appropriate stress is helpful to regulate bone remodeling during tooth movement, to provide new methods and ideas for accelerating orthodontic tooth movement, which has clinical guiding significance.
Material and methods
Ethics statement
The Ethics Committee of Shanghai Stomatological Hospital (China) has granted the study protocol, and informed consent was got from the parents of three donors (one 12-year-old female and two 13-year-old males) following the Declaration of Helsinki.
Cell culture and treatment
HPDLSCs were removed from the root surface of healthy premolars extracted following orthodontic extraction. Culture methods were applied as reported in the literature [15]. Cells between passages 4 and 8 were used for experiments.
Mouse bone marrow mesenchymal stem cells (mBMSCs) were obtained from the tibias and femurs of 4-week-old wild-type mice and TRPC6−/− mice. Culture methods were applied as reported in the literature [17]. When mBMSCs were 80% confluent, they were passaged for all experiments.
Cyclic strain experiments
HPDLSCs were cultured on six-well Bioflex plates (Flexcell International, Hillsborough, NC, USA) at a density of 3*105 cells/well. At 90% confluence, cells were serum-starved in α-MEM for 24 h and were then subjected to cyclic strain using a Flexcell Tension Plus system (FX-5000 T, Flexcell International, USA) (10%, 0.1 Hz) for 6 or 24 h. As static controls, cells were seeded into the same kind of plates but were not subjected to cyclic strain.
3D scaffold preparation
Cells at a density of 4*107 cells/ml were mixed with an equal volume of 4% low melting point agarose (Sigma, Merck KGaA, Darmstadt, Germany). The mixture was added to 6-well BiopressTM compression culture plates (Flexcell International, Burlington, NC, USA) and allowed to solidify at room temperature.
Cyclic compression experiment
Cells within the 3D scaffolds were incubated in α-MEM for 24 h and then subjected to cyclic strain using a Flexcell-5000C™ compression system (Flexcell International, Burlington, NC, USA) (2 kPa, 0.1 Hz) for 6 and 24 h [18,19,20,21]. Unloaded 3D scaffold samples were used as static controls.
Fabrication of confinement slide with different height
The new silicon wafer (Yangzhou Genesis Microelectronics Co., Ltd, China) was washed with isopropanol and dried with gentle nitrogen. And then silicon wafer was placed on a heating plate at 120 °C for 30 min. After recovery to room temperature, the silicon wafer was coated with an SU-8 photoresist according to the instructions to obtain different heights by a spin coater (SPIN200-IND, Germany). And the soft-baked temperature was set up following the SU-8 photoresist instructions. The prepared silicon was covered by a patterned mask which was designed before this experiment and exposed under the mask aligner (URE-2000S/A, China). Sequentially, the silicon wafer was immersed in the developer completely. After rinsing the silicon wafer with isopropanol to remove the developer completely, the silicon was prepared for fabricating different height confinement slides (3 μm, 5 μm, 8 μm, 10 μm). The height of the silicon wafer was measured by an optical profilometer (Alphastep D-600). The glass slides (diameter, 12 mm, Matsunami, Japan) were cleaned with plasma cleaner (Harrick PDC-002, USA) for 3 min before using. The silicon wafer was covered with RTV615 PDMS (Momentive, USA) mixture (A/B, 8/1, w/w), and then the glass slide was gently pressed on PDMS. After heating at 120 °C for 15 min, the confinement slide containing PDMS pillars was prepared.
Construction of cell confined microenvironment in vitro
The experiment methods were applied according to the reported literature [22]. Briefly, the large PDMS pillars (A/B, 35/1, w/w) were stuck on the lid of the 6-well glass bottom plate (Matsunami, Japan), which were made before using the custom mold. The PDMS pillars on the glass slide were modified with 0.1 mg/mL PLL(20)-g [3.5]-PEG(2) (10 mM HEPES, pH 8.5) (SuSoS, Switzerland) at room temperature for at least 3 h. After rinsing with PBS (10 mM, pH 7.4), the glass slides containing PDMS pillars were stuck on the other side of the large PDMS pillars.
The hPDLSCs or mBMSCs were seeded into a 6-well glass bottom plate one day in advance. The cells were stained with SiR-actin (200 nM) for 6 h and Hoechst (200 ng/mL) for 1 h at least. To inhibit the activation of TRPC6, SKF96365 (100 µM) (Selleck, USA) was added to the medium for 1 h before incubation in the confined microenvironment [23]. The prepared lid of the 6-well glass bottom plate was vertically covered on the cells and maintained with adhesive tape (Scotch Magic Tape, 3 M).
Live-cell imaging
The motility of live-cell under confinement microenvironments was acquired by an inverted fluorescence microscope with 20 × objective (Leica, Germany). All cells were stained using Hoechst (cell nucleus) and SiR-actin (cell actin cytoskeleton). The microscope was equipped with a humidified incubation chamber at 37 °C and 5% CO2 at all times. MetaMorph software (Universal Imaging) was used to analyze cell migration images.
Image analysis
Cells trajectories were tracked from fluorescence images of Hoechst staining with ImageJ (using the TrackMate or manual tracking with the TrackMate plugin) and a home-written Matlab program. We calculated cells speed according to the reported literature [22].
Cell cytoskeleton analyses were obtained from fluorescence images of SiR-actin and analyzed by ImageJ software.
Construction of transcriptome libraries and sequencing
Total RNA was extracted from hPDLSCs after application of tensile force and pressure force in vitro using TRI reagent (Sigma-Aldrich, Germany). RNA-seq libraries were generated using these RNA sequences by RNA fragmentation, cDNA synthesis with random hexamer primers, linker ligation and amplification. Illumina HiSeq 2000 platform was used to sequence each library.
RNA-seq data analysis
For gene expression annotation, the RPKM (reads per kb per million mapped reads) value of each gene was counted, and DEGs were defined as those with a fold change of ≥ 2.5 and a P value of ≤ 0.01. For analysis of expression patterns, hierarchical clustering, functional enrichment and protein–protein interaction analyses were conducted with the omicsbean website (http://www.omicsbean.cn) using the DAVID Gene Ontology database.
Quantitative reverse transcription-polymerase chain reaction (PCR)
Total RNA was extracted from hPDLSCs in the confined microenvironment and from mBMSCs isolated from wild-type mice and TRPC6−/− mice, and reverse transcription and real-time PCR were performed as reported in the literature [24]. The primers were designed using Primer Premier 5.0 software and are listed in Table 1.
Western blot analysis and antibodies
HPDLSCs after application of tensile force, pressure force were scraped with ice-cold RIPA buffer (Thermo Fisher Scientific, USA). The method of western blot was applied according to the reported literature [15]. Membranes were incubated overnight at 4 ℃ with the following primary antibodies: anti-TRPC4 (Sigma, MO); anti-TRPC6 (Abcam, UK) and anti-β-actin (Sigma, MO). ImageJ software was used to analyze the relative density of each band.
Animal models and orthodontic force application
All protocols were approved and conducted in accordance with the regulations of the Institutional Animal Care and Use Committee of Shanghai Model Organisms Center (China). 24 Sprague–Dawley rats and 48 male mice (24 wild-type mice and 24 TRPC6−/− mice) were used. TRPC6−/− mice were constructed at the Shanghai Model Organisms Center using CRISPR/Cas9 genome editing (Additional file 1: Fig. S1) [15].
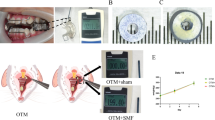
To establish the OTM model in rats, 24 Sprague–Dawley rats (male, 8 weeks old) weighing 200 g were used: 6 rats per day on Days 1, 3, 7 and 14, with at least two rats per day for micro-CT and immunohistochemical staining. To generate tooth movement, a nickel-titanium coil spring (Smart Technology, China) was used to connect the maxillary right first molar to the incisors (actuation force = 60 g). The left first molar was used as the control.
To establish the OTM model in mice, 24 wild-type mice and 24 TRPC6−/− mice (male, 4 weeks old) weighing 20 g were used: 6 mice per day on Days 1, 3, 7 and 10, with at least two mice per day for micro-CT and immunohistochemical staining. The method of appropriate mechanical force application was as described above, but the actuation force was 20 g.
OTM distance measurement and micro-CT analysis
Using an overdose of pentobarbital sodium, we sacrificed the rats and mice and harvested the maxillae. The OTM distance refers to the distance between the first molar and the second molar. Then, we used a high-resolution micro-CT system (Scanco Medical, Switzerland) to image the maxillae. We chose the furcation of the first molar as the location for analysis of bone mineral density.
Immunohistochemical staining
Immunohistochemical staining was conducted as previously described [25]. Anti-TRPC6 (ab62461; Abcam, Cambridge, UK) and anti-COL1 (ab34710; Abcam, Cambridge, UK) primary antibodies were used.
TRAP staining
We performed TRAP staining with an acid phosphatase kit (Sigma, USA) according to the manufacturer’s instructions as previously described [25]. We chose 3–5 randomly selected fields near the PDL for counting of TRAP-positive cells.
Statistical analysis
The experimental data are shown as the means ± standard deviations, and each experiment was repeated three times independently. Statistical analyses and mapping were performed with GraphPad Prism 8. For multiple groups of samples, if the data obeyed the normal distribution and the variance was homogeneous, ANOVA was used, and the Bonferroni test was used for pairwise comparisons; if the data obeyed the normal distribution but the variance was nonhomogeneous, Welch test was used, and Dunnett’s T3 test was used for pairwise comparisons; if the data were nonnormally distributed, the Kruskal–Wallis test with the X2 statistic was adopted, and the Bonferroni test was used for pairwise comparisons. P < 0.05 was considered to indicate a significant difference.
Results
The rapid identification of the important stress-sensing gene TRPC6 by RNA-seq
We investigated hPDLSCs after the application of tensile and compressive forces in vitro. Principal component analysis (PCA) was performed after the application of the tensile and compressive forces (Additional file 1: Fig. S2A, B). We identified 7091, 7722, 3302 and 6433 differentially expressed genes(DEGs) in the cells compared with the control cells after tensile force application for 6 h, tensile force application for 24 h, compressive force application for 6 h and compressive force application for 24 h, respectively. In the tensile force groups, we identified 4038 significantly upregulated DEGs and 3053 significantly downregulated DEGs between 6 h and control groups, as well as 3855 significantly upregulated DEGs and 3867 significantly downregulated DEGs between the 24 h and control groups (false discovery rate (FDR) < 5%, > twofold change). In the compressive force groups, we identified 1508 significantly upregulated DEGs and 1794 significantly downregulated DEGs between 6 h and control groups, as well as 2832 significantly upregulated DEGs and 3601 significantly downregulated DEGs between the 24 h and control groups (false discovery rate (FDR) < 5%, > twofold change), which formed distinct clusters (Additional file 1: Fig. S2C, D). The relative differential expression values for the different times are directly indicated on the heatmap (Additional file 1: Fig. S2E, F). According to PPI network analysis, we found several significant DEGs, such as TRPC1, TRPC4, TRPC6, CUL5, PRKCA, BMP4, PLCB3, PLCB4, MYC, ROCK2, CD19, WNT16 and Wnt11 (Fig. 1A, B). Due to our previous studies on calcium channels and mechanosensitive channels [26, 27], we focused on the TRPC channel, such as TRPC4 and TRPC6.
TRPC6 regulated the stress response of periodontal ligament stem cells
To validate the RNA-seq results in vitro, we used western blotting to evaluate TRPC4 and TRPC6 protein expression in periodontal ligament stem cells after stress in vitro. HPDLSCs showed time-dependent activation of TRPC6 (Fig. 2A) and TRPC4 (Additional file 1: Fig. S3A) under both tensile and compressive stress. The change in TRPC6 was more obvious after compressive stimulation than after tensile stimulation. The change in the level of TRPC6 was greater than that in the level of TRPC4; thus, we mainly studied TRPC6.
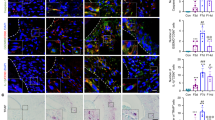
Mechanical force upregulated TRPC6 expression in PDLSCs. A The expression of TRPC6 as determined by western blotting and quantitative analysis of TRPC6 expression in hPDLSCs are shown. B The immunohistochemical images indicate that the expression of TRPC6 increased in molars subjected to orthodontic force. The black arrow indicates the direction of orthodontic force. C, crown; R, root; P, periodontal ligament; A, alveolar bone; Black scale bar, 1 mm; White scale bar, 500 μm; Blue scale bar, 200 μm. C Schematic diagrams. D Semiquantitative analysis of TRPC6-positive cells in immunohistochemical images. E The results of real-time PCR showed that the expression of TRPC6 was significantly increased when hPDLSCs were exposed to a 3 μm confined microenvironment for 24 h. All the results are expressed as the mean ± standard deviation of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns, P > 0.05
In vivo, after force application, the proportions of TRPC6-positive cells on Days 3 and 7 were increased more than those in the control group (Fig. 2B-D). In addition, the proportions of TRPC4-positive cells on Days 3 and 7 were increased compared with those in the control group (Additional file 1: Fig. S3B). These results showed that TRPC4 and TRPC6, as sensors of mechanical force, were activated in PDLSCs after appropriate mechanical stimulation.
In addition, real-time PCR revealed that TRPC6 expression was significantly increased in hPDLSCs in a 3 μm confined microenvironment (Fig. 2E), further verifying the mechanosensitivity of TRPC6.
Inhibition of TRPC6 changed the morphology and migration speed of hPDLSCs in the confined microenvironment
We induced confinement by covering cells with an adhesive surface as previously described [21]. The initial height of the hPDLSCs was approximately 10 μm. At a height of 8 μm (slight restriction), the cell body was slightly deformed. At heights of 5 μm and 3 μm (high restriction), both the cell body and the nucleus were severely deformed [22, 28].
In the 5 μm confined microenvironment, the actin bundles in the cytoskeleton disappeared, and the cell body became round and disordered. Under treatment with the TRPC6 inhibitor SKF96365, the ability of cells to resist compressive stimulation decreased; thus, the cell morphology exhibited a relaxed state (Fig. 3A). In addition, we found that nuclear restriction (5 μm) decreased the speed of cell migration. Environmental factors played a key regulatory role, while the role of TRPC6 was not obvious (Fig. 3B).
HPDLSCs in the 5 μm and 8 μm confined microenvironments. A In the 5 μm confined microenvironment, the actin bundles in the cytoskeleton disappeared, and the cell body became round and disordered. In the 8 μm confined microenvironment, the changes in the cytoskeleton were not obvious. Under treatment with the TRPC6 inhibitor SKF96365, the ability of cells to resist compressive stimulation decreased; thus, the cell morphology exhibited a relaxed state. Scale bar, 20 μm. B The cell migration trajectory decreased in the confined microenvironment. Environmental factors played a key regulatory role, and the role of TRPC6 was also significant in the 8 μm confined microenvironment. The results are expressed as the mean ± standard deviation of one independent experiment; N ≥ 3. *P < 0.05; ***P < 0.001; ns, P > 0.05
In the 8 μm confined microenvironment, the changes in the cytoskeleton were not obvious. The cell migration trajectory decreased, while the TRPC6 inhibitor SKF96365 also decreased the speed of cell migration. Environmental factors played a regulatory role, while the role of TRPC6 was also significant (Fig. 3B).
TRPC6 knockout changed the morphology and migration speed of mBMSCs in the confined microenvironment
MBMSCs were obtained from wild-type mice and TRPC6 KO mice. The height of the mBMSCs was approximately 12 μm; thus, we chose to limit our investigation to heights of 5 μm and 10 μm. At a height of 10 μm (slight restriction), the cell body was slightly deformed. At a height of 5 μm (high restriction), both the cell body and the nucleus were severely deformed.
In the 5 μm confined microenvironment, mBMSCs resisted compressive stimulation, which made the cells acquire a rounded shape. In the 10 μm confined microenvironment, the changes in the cytoskeleton were not obvious. With TRPC6 knockout, the ability of cells to resist compressive stimulation decreased; thus, the cell morphology exhibited a relaxed state (Fig. 4A), while the cell migration trajectory decreased, and TRPC6 knockout also decreased the speed of cell migration in the 10 μm confined microenvironment (Fig. 4B). Environmental factors did not play a regulatory role, while the role of TRPC6 was significant. However, when the restriction was severe at 5 μm, environmental factors played a leading role, and the influence of the genetic factor TRPC6 was not obvious (Fig. 4B).
MBMSCs in the 5 μm and 10 μm confined microenvironments. A In the 5 μm confined microenvironment, mBMSCs resisted compressive stimulation, which made the cells acquire a rounded shape. In the 10 μm confined microenvironment, the changes in the cytoskeleton were not obvious. With TRPC6 knockout, the ability of cells to resist compressive stimulation decreased; thus, the cell morphology exhibited a relaxed state. Scale bar, 20 μm. B The cell migration trajectory decreased with TRPC6 knockout, and TRPC6 knockout decreased the speed of cell migration in the 10 μm confined microenvironment. The results are expressed as the mean ± standard deviation of one independent experiment; N ≥ 2. **P < 0.01; ***P < 0.001; ns, P > 0.05
TRPC6 knockout suppressed osteoblast and osteoclast differentiation, leading to a shorter OTM distance
To test whether TRPC6 plays a significant role in periodontal tissue remodeling, we used TRPC6−/− mice (TRPC6 KO mice) to prove the effect of TRPC6. We performed micro-CT scanning of the furcation region of the first maxillary molar, and we confirmed that TRPC6 KO mice showed a decreased OTM distance and an increased BMD (indicating slower bone mineral density conversion) compared to those in wild-type mice (Fig. 5A, B).
TRPC6 knockout suppressed osteoblast and osteoclast differentiation, leading to a shorter OTM distance. A TRPC6 knockout led to a shorter OTM distance. Scale bar, 1 mm. B TRPC6 knockout led to slower bone mineral density conversion. C TRPC6 knockout led to decreases in TRPC6 and COL1 expression and TRAP-positive osteoclasts. The black arrow indicates the direction of orthodontic force. Scale bar, 500 μm. Semiquantitative analysis of TRPC6 expression, COL1 expression and TRAP-positive osteoclasts. The results are expressed as the mean ± standard deviation of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
On Day 7, histopathological examination of mice subjected to strain revealed that TRPC6 KO mice showed lower TRPC6 staining and COL1 staining than wild-type mice (Fig. 5C). Consistent with this finding, we found a decrease in the percentage of TRAP-positive osteoclasts on the compression side of the alveolar bone in TRPC6 KO mice compared to control mice (Fig. 5C). These in vivo findings collectively confirmed that the mechanosensitivity of TRPC6 is important in periodontal tissue remodeling.
Discussion
TRPC6 is a nonselective cation channel. In our study, we showed that TRPC6 mediated the mechanical response of PDLSCs in an OTM model and cultured hPDLSCs subjected to tensile and compressive forces, contributing to periodontal tissue remodeling. First, the important stress-sensing gene TRPC6 was rapidly identified by RNA-seq; second, upregulation of TRPC6 in PDLSCs was induced by appropriate mechanical force application both in vivo and in vitro; and third, the abovementioned hypothesis was further verified in confined microenvironments; finally, reductions in osteogenic and osteoclastic activities and tooth movement distances were observed in the OTM model of TRPC6 knockout mice.
PDLSCs are mechanosensitive and are responsive to mechanical stimulation, which endows them with an important role in periodontal tissue remodeling [29,30,31,32]. However, the mechanism by which PDLSCs are involved in mechanosensing in periodontal tissue is unclear. In-depth study of gene expression profiles is complex, and RNA sequencing is one of the best methods for this type of research at present. HPDLSCs in the presence or absence of force loading with the Flexcell loading system in vitro were analyzed. By RNA-seq, we found that TRPC4 and TRPC6 were important in the transduction of mechanical stimuli in PDLSCs. TRPCs can be classified into four subsets according to their amino acid similarity: TRPC1, TRPC2, TRPC3/6/7 and TRPC4/5 [33]. Despite their differences, both TRPC4 and TRPC6 have been implicated as channels that may be sensitive to stress. TRPC4 and TRPC6 are expressed in various types of smooth muscle cells [34, 35]. Static strain increases TRPC4 mRNA expression in myometrial smooth muscle, and TRPC6 knockdown in cerebral arteries inhibits pressure-induced depolarization and contraction [36, 37]. Based on the changes in the levels of TRPC6 and TRPC4 and our previous research, we demonstrated the regulatory effect of mainly TRPC6 under stress in vivo and in vitro.
In the study of mechanical force, we should not only consider whether the force is applied by tension or compression but also pay attention to the value. The force on the cells should not be too large; otherwise, the cells detach and die. The force should also not be too small. If it is too small, the mechanical signal may not be activated. Therefore, the “optimum force value” has also become a focus of attention. For tension, it is generally considered that 10–15% is the appropriate value and that values greater than or equal to 20% are excessive [38]. For compression, it is generally considered that the maximum value is 5 kPa [18,19,20,21]. Therefore, the appropriate forces used in our study were a tensile force of 10% and a compressive force of 2 kPa. In vivo, application of the appropriate force does not lead to adverse effects such as root resorption and alveolar bone resorption. The best force value can result in a faster tooth movement speed. Therefore, an actuation force of 60 g was used in rats, and an actuation force of 20 g was used in mice in our study [39].
This study is the first to prove that TRPC6 is a mechanosensitive gene in periodontal tissue, providing novel insight into mechanosignaling. The confined microenvironment is the highlight and innovation of this study. In the process of orthodontic tooth movement, cells on the pressure side are under pressure, and there is also mutual extrusion between the cells, resulting in a confined microenvironment of the cells. The in vivo model and the Flexcell system cannot be used to visually observe the changes in a single cell under stress, but the confined microenvironment generated by microfluidics combined with microscopy can be used to observe the dynamic changes in a single cell under stress at the micron level. Real-time imaging is not possible in the in vivo model and Flexcell system, but the confined microenvironment we generated can provide this ability. Mesenchymal stem cells experience mechanical restriction of the cell body and nucleus in the confined microenvironment, but the effect of this mechanical signal on the morphology, migration and function of mesenchymal cells is not clear. The changes induced in the cytoskeleton and migration of cells in the confined microenvironment are similar to those induced under pressure, as shown in the referenced studies [40,41,42]. Therefore, the change in the migration velocity of periodontal ligament stem cells in a confined microenvironment leads to a change in the tooth movement velocity under the same conditions produced by TRPC6 knockout.
Through the discovery of TRPC6 and the related regulatory mechanisms, we found that TRPC6 plays an important regulatory role under both stress and confined microenvironment conditions. We have not conducted a detailed study on the different regulatory mechanisms of TRPC6. We expect to find a unified and important molecular regulatory mechanism of TRPC6. Therefore, we generated a confined microenvironment and observed the changes in the cells in the confined microenvironment at the micron level to confirm the regulatory role of TRPC6.
Gene knockout studies can more effectively explain the roles of related genes in regulating the mechanism of periodontal tissue remodeling and provide more reliable evidence. Therefore, in this study, CRISPR/Cas9 genome editing was used to introduce mutations via nonhomologous recombination repair, resulting in a reading frame shift and functional loss of the TRPC6 gene. In our study, TRPC6 knockout suppressed osteoblast and osteoclast differentiation, leading to a shorter OTM distance, verifying the mechanosensitivity of TRPC6.
The identity of TRPC6 is a mechanically controlled cation channel has been confirmed by many studies [13, 14]. However, we found that the function of TRPC6 may not be limited to acting as an ion channel; it may also function as a cell membrane receptor that can transmit mechanical signals. The specific signal transmission mechanism is unclear; however, TRPC6 expression may change in response to mechanical stimuli, and TRPC6 expression increases after application of appropriate stress.
Conclusion
In conclusion, PDLSCs modulate periodontal tissue remodeling in response to appropriate mechanical stimulation through TRPC6; however, when the stress is excessive, alveolar bone and tooth roots are readily absorbed. Under this condition, environmental factors play a leading role, and the regulatory effect of TRPC6 is not obvious. Further studies on the specific mechanism of TRPC6, experiments on mechanically controlled Ca2+ influx and studies in selected knockout mice should be performed to prove the regulatory role of TRPC6 in alveolar bone remodeling. In addition, we should devote more attention to the clinical application of TRPC6 and the important roles of small molecule compounds such as TRPC6 inhibitors and activators.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- OTM:
-
Orthodontic tooth movement
- PDLSCs:
-
Periodontal ligament stem cells
- hPDLSCs:
-
Human PDLSCs
- BMD:
-
Bone marrow density
- RNA-seq:
-
RNA sequencing
- mBMSCs:
-
Mouse bone marrow mesenchymal stem cells
- MICs:
-
Mechanosensitive ion channels
- ENaCs:
-
Epithelial sodium channels
- K2Ps:
-
Two-pore potassium channels
- TRPs:
-
Transient receptor potential channels
- Piezos:
-
Piezo electric ion channels
- TRPC6:
-
Transient receptor potential channel 6
- PCA:
-
Principal component analysis
- DEGs:
-
Differentially expressed genes
- FDR:
-
False discovery rate
References
Le HQ, Ghatak S, Yeung C, Tellkamp F, Wickstrm SAA. Mechanical regulation of transcription controls polycomb-mediated gene silencing during lineagecommitment. Nat Cell Biol. 2016;18(8):864–75.
Zhang L, Issa Bhaloo S, Chen T, et al. Role of resident stem cells in vessel formation and arteriosclerosis. Circ Res A J Am Heart Assoc. 2018;122(11):1608–24.
Shi Y, Wang Y, Li Q, Liu K, Hou J, Shao C, et al. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat Rev Nephrol. 2018;14(8):493–507.
Volponi AA, Pang Y, Sharpe PT. Stem cell-based biological tooth repair and regeneration. Trends Cell Biol. 2010;20(12):715–22.
Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366(9499):1809–20.
Meikle MC. The tissue, cellular, and molecular regulation of orthodontic tooth movement: 100 years after Carl Sandstedt. Eur J Orthod. 2006;28(3):221–40.
Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364(9429):149–55.
Chalfie M, Chalfie M. Neurosensory mechanotransduction. Nat Rev Mol Cell Biol. 2009;10:44–52.
Douguet D, Honoré E. Mammalian mechanoelectrical transduction: structure and function of force-gated ion channels. Cell. 2019;179(2):340–54.
Nilius B, Owsianik G. The transient receptor potential family of ion channels. Genome Biol. 2011;12(3):218.
Clapham DE. Calcium signaling. Cell. 2007;131(6):1047–58.
Yang J, Tang L, Zhang F, Yang T, Lu T, et al. Sevoflurane preconditioning promotes mesenchymal stem cells to relieve myocardial ischemia/ reperfusion injury via TRPC6-induced angiogenesis. Stem Cell Res Ther. 2021;12(1):584.
Nikolova-Krstevski V, Wagner S, Yu ZY, Cox CD, Cvetkovska J, et al. Endocardial TRPC-6 channels act as atrial mechanosensors and load-dependent modulators of endocardial/myocardial cross-talk. JACC Basic Transl Sci. 2017;2(5):575–90.
Dalrymple A, Mahn K, Poston L, Songu-Mize E, Tribe RM. Mechanical stretch regulates TRPC expression and calcium entry in human myometrial smooth muscle cells. Mol Hum Reprod. 2007;13(3):171–9.
Li W, Jing M, Bsa B, Gang Y, Sl B, Mcb D, Lya B, Jie P, Yla B. Role of transient receptor potential channel 6 in the osteogenesis of periodontal ligament stem cells. Int Immunopharmacol. 2021;100:108134.
Ma N, Adnot S, Archer SL, Dupuis J, Lloyd JP, Maclean MR, et al. Cellular and molecular basis of pulmonary arterial hypertension. J Am Coll Cardiol. 2009;54(1):S20–31.
Sun H, Wu Y, Fu D, Liu Y, Huang C. SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-κB signaling pathway. Stem Cells. 2014;32(7):1943–55.
Strobl H, Krump C, Borek I. Micro-environmental signals directing human epidermal Langerhans cell differentiation. Semin Cell Dev Biol. 2019;86:36–43.
Sanchez C, Pesesse L, Gabay O, Delcour JP, Msika P, Baudouin C, et al. Regulation of subchondral bone osteoblast metabolism by cyclic compression. Arthritis Rheum. 2014;64(4):1193–203.
Sanchez C, Gabay O, Salvat C, Henrotin YE, Berenbaum F. Mechanical loading highly increases IL-6 production and decreases OPG expression by osteoblasts. Osteoarthr Cartil. 2009;17(4):473–81.
Gabay O, Hall DJ, Berenbaum F, Henrotin Y, Sanchez C. Osteoarthritis and obesity: experimental models. Joint Bone Spine. 2008;75(6):675–9.
Liu YJ, Berre ML, Lautenschlaeger F, Maiuri P, Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160(4):659–72.
Rachubik P, Szrejder M, Rogacka D, Audzeyenka I, Piwkowska A. The TRPC6-AMPK pathway is involved in cytoskeleton reorganization and glucose uptake in podocytes. Cell Physiol Biochem. 2018;51(1):393–410.
Jin SS, He DQ, Wang Y, Zhang T, Yu HJ, Li ZX, Zhu LS, Zhou YH, Liu Y. Mechanical force modulates periodontal ligament stem cell characteristics during bone remodeling via TRPV4. Cell Prolif. 2020;53(10):e12912.
Chen MH, Wang YH, Sun BJ, Yu LM, Chen QQ, Han XX, Liu YH. HIF-1α activator DMOG inhibits alveolar bone resorption in murine periodontitis by regulating macrophage polarization. Int Immunopharmacol. 2021;99:107901.
Ju Y, Ren X, Zhao S. Distal C-terminus of Cav1.2 is indispensable for the chondrogenic differentiation of rat dental pulp stem cells. Cell Biol Int. 2020;44(2):512–23.
Gao Q, Ge J, Ju Y, Li C, Gao J, Wu M, et al. Roles of L-type calcium channels (CaV1.2) and the distal C-terminus (DCT) in differentiation and mineralization of rat dental apical papilla stem cells (rSCAPs). Arch Oral Biol. 2017;74:75–81.
Maël LB, Johannes A, Matthieu P. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol. 2012;4(11):1406–14.
Feng L, Yang R, Liu D, Wang X, Song Y, Cao H, et al. PDL progenitor-mediated PDL recovery contributes to orthodontic relapse. J Dent Res. 2016;95(9):1049–56.
Tao S, Qiu L, Chang H, Yang Y, Dong S. Cyclic tension promotes osteogenic differentiation in human periodontal ligament stem cells. Int J Clin Exp Pathol. 2014;7(11):7872–80.
Yamaguchi M, Shimizu N. Identification of factors mediating the decrease of alkaline phosphatase activity caused by tension-force in periodontal ligament cells. Gen Pharmacol. 1994;25(6):1229–35.
Li Q, Yang G, Li J, Ding M, Zhou N, Dong H, Mou Y. Stem cell therapies for periodontal tissue regeneration: a network meta-analysis of preclinical studies. Stem Cell Res Ther. 2020;11(1):427.
Hamill OP, Maroto R. TRPCs as MS channels. Curr Top Membr. 2007;59:191–231.
Beech DJ. Emerging functions of 10 types of TRP cationic channel in vascular smooth muscle. Clin Exp Pharmacol Physiol. 2005;32(8):597–603.
Dietrich A, Chubanov V, Kalwa H, Rost BR, Gudermann T. Cation channels of the transient receptor potential superfamily: their role in physiological and pathophysiological processes of smooth muscle cells. Pharmacol Ther. 2006;112(3):744–60.
Dalrymple A, Mahn K, Poston L, Songu-Mize E, Tribe RM. Mechanical stretch regulates TRPC expression and calcium entry in human myometrial smooth muscle cells. Mol Hum Reprod. 2007;13(3):31–9.
Welsh D, Morielli A, Nelson M, Brayden J. Transient receptor potential channels regulate myogenic tone of resistance arteries. Circ Res. 2002;90(3):248–50.
King GJ, Keeling SD, McCoy EA, et al. Measuring dental drift and orthodontic tooth movement in response to various initial forces in adult rats. Am J Orthod Deniofac Orthop. 1991;99(5):456–65.
Mussig E, Schulz S, Spatz JP, Ziegler N, Tomakidi P, Steinberg T. Soft micropillar interfaces of distinct biomechanics govern behaviour of periodontal cells. Eur J Cell Biol. 2010;89(4):315–25.
Doolin MT, Stroka KM. Physical confinement alters cytoskeletal contributions towards human mesenchymal stem cell migration. Cytoskeleton (Hoboken). 2018;75(3):103–17.
de Araujo RM, Oba Y, Kuroda S, Tanaka E, Moriyama K. RhoE regulates actin cytoskeleton organization in human periodontal ligament cells under mechanical stress. Arch Oral Biol. 2014;59:187–92.
McGregor AL, Hsia CR, Lammerding J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr Opin Cell Biol. 2016;40:32–40.
Acknowledgments
We acknowledge Professor Hongxiu Yu and Tingjiao Liu for providing important research suggestions in this study.
Funding
This work was supported by National Natural Science Foundation, China (Nos. 81771109, 82071153 and 31870978); Shanghai Municipal Commission of Science and Technology, China (No. 22ZR1454200); Shanghai Stomatological Hospital Talent Project, China (No. SSDC-2019-RC01).
Author information
Authors and Affiliations
Contributions
LW performed most experiments and wrote the manuscript. HL performed the experiments under the confined microenvironment and revised the manuscript. BS and XT assisted in completing cell experiments. JM, YW and MC assisted in completing animal experiments. LY and JP collected data and performed analyses. SL, Y-JL and YL designed experiments and revised the article. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Ethics Committee of Shanghai Stomatological Hospital (China) has granted the study protocol, and informed consent was got from the parents of three donors (one 12-year-old female and two 13-year-old males) following the Declaration of Helsinki (No. 2019-003). All protocols were granted and conducted following the regulations of the Institutional Animal Care and Use Committee of Shanghai Model Organisms Center (China) (No. 2021-0005).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1
. The generation principle and identification of TRPC6 knockout mice. (A) The generation principle of TRPC6 knockout mice. (B) The scheme for identifying the genotype of TRPC6 knockout mice (C) The results of real-time PCR showed that the expression of TRPC6 was knocked out in mBMSCs from TRPC6 knockout mice. Fig. S2. PCA, cluster and heatmap analyses under stress conditions. (A) PCA under tension. (B) PCA under compression. (C) Cluster analysis under tension. (D) Cluster analysis under compression. (E) Heatmap analysis under tension. (F) Heatmap analysis under compression. Fig. S3. Mechanical force upregulated TRPC4 expression in periodontal ligament stem cells. (A) The expression of TRPC4 as determined by western blotting and quantitative analysis of TRPC6 expression in hPDLSCs are shown. (B) The expression of TRPC4 increased after orthodontic force application in rats; a representative image of TRPC4 immunohistochemical staining on the tension and compression sides of molars subjected to orthodontic force is shown. The black arrow indicates the direction of orthodontic force. Black scale bar, 1 mm; White scale bar, 500 μm; Blue scale bar, 200 μm; Semiquantitative analysis of TRPC4-positive cells. The results are expressed as the mean ± standard deviation of three independent experiments. *P < 0.05; ***P < 0.001.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, L., Liang, H., Sun, B. et al. Role of TRPC6 in periodontal tissue reconstruction mediated by appropriate stress. Stem Cell Res Ther 13, 401 (2022). https://doi.org/10.1186/s13287-022-03055-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-022-03055-z