Abstract
Background
Due to indels in the β-globin gene, patients with β-thalassemia major exhibit a range of severity, with genotype β0β0 > β0β+ > β+β+, according to the production level of the β-globin chain. More than 300 mutations have been identified in the β-globin gene.
Case presentation
In this case study, we report a compound heterozygous condition with a rare concoction of four different variants (CD 3(T > C), CD41/42 (-CTTT), IVS II-16 (G > C), and IVS II-666 (C > T) in a single β-globin gene. A regular transfusion-dependent 4-year-old male patient from India was included in the study. Augmented direct sequencing of the β-globin gene helped reveal the presence of an unusual combination of different variants in a single gene. This patient clinically presented as β-thalassemia major and was genotypically considered as β0β+, although CD41/42(-CTTT) was the only causative/pathogenic mutation in the disease severity.
Conclusion
Although CD41/42-(CTTT) is the only pathogenic variant among the four variants, the clinical complications of such a combination of variants (pathogenic and benign) is not well understood. Intronic mutations may have the ability to modify clinical characteristics. The variants must therefore be reclassified using additional mRNA splicing and expression-based studies. Additionally, these types of combinations may have significance in studying population migration around the world.
Similar content being viewed by others
Background
Tetrameric hemoglobin molecules are involved in gaseous exchange in red blood cells. The β-globin chains, when paired with the α-globin chains, result in the formation of hemoglobin tetramers. It has come to light that mutations in the β-globin gene cause either insufficient (β+) or halted development (β0), resulting in β-thalassemia major. The possible genotypes responsible for β-thalassemia major, in order of severity, are β0β0 > β0β+ > β+β+ 1.
Various studies have identified more than 300 mutations to date (Ithanet and HbVar databases). The Mediterranean region, the Indian subcontinent, Southeast Asia, and the Middle East are all home to these mutations 2, and thousands of people of various ethnicities are affected by this autosomal recessive syndrome 3. The advent of direct or Sanger DNA sequencing has increased the study of mutations in various diseases and β-thalassemia. Here, we present a case of a rare compound heterozygous condition involving four variants in the β-globin gene identified by direct sequencing. We discovered this patient with four separate variants in the β-globin gene (CD 3 [T>C], CD 41/42 [-CTTT], IVS II-16 [G>C], and IVS II-666 [C>T]) during a screening of variants among β-thalassemia major patients registered at the JNMC hospital, Aligarh, India.
Case presentation
A 4-year-old Indian boy from a low-income family (farmer) was admitted to the hospital for a routine blood transfusion. His weight and height was between the 10th and 50th centile, with −1.71 and −1.54 Z-scores, respectively. He had O (+ve) blood group and required 10–12 blood transfusions a year. His adult hemoglobin (HbA2) was > 3%, mean corpuscular volume (MCV) was 74.89 fl, and mean corpuscular hemoglobin (MCH) was 30.4 pg according to his medical records, and his physical examination revealed that he had knee pain and an enlarged spleen (> 3 cm below costal margin). As a result, clinicians discovered that he had β-thalassemia major and he was on folic acid 2.5 mg/day and deferasirox 30 mg/kg/day. After reviewing his family history, it was discovered that one of his two male siblings (who died) had β-thalassemia, while all of his sisters were well (Fig. 1). The parents gave their consent for the patient, but the parents and their healthy offspring declined to participate, so we removed them from the study. Medical details and blood samples were collected from the hospital once written informed consent was given. The genomic DNA was isolated according to the manufacturer’s instructions using the gDNA Blood Mini Kit (Chromous Biotech, Bengaluru, India). Additional genomic analysis was carried out according to [4]. The BLAST method in the National Center for Biotechnology Information (NCBI) database was used to confirm the identification of the collected sequences. By comparing mutations to reference sequences, direct counting was used to determine their presence (NG 000007.3 Homo sapiens chromosome 11). The dbSNP, Clinvar, Ithanet, and HbVar databases were then used to look for mutations that had been identified in other populations [5].
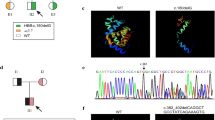
Details related to the case study. A The pedigree diagram illustrates family history: one male sibling (deceased) was affected with β-thalassemia major and other siblings were unaffected. B Sequencing data shows wild type and mutant chromatograms for both exonic (CD3 [T>C] and CD41/42 [-CTTT]), and intronic (IVS II-16 [G>C] and IVS II-666 [C>T]) variants. C Clinical data collected from the hospital. D Schematic diagram of HBB gene structure with four identified variants at their respective locations
Discussion
The patient, from western Uttar Pradesh in India, has a unique combination of four different mutations in a single gene (β-globin), along with typical developmental milestones, although he had a lower body mass index (BMI) (10–50th centile, −1.71 Z-score), splenomegaly, and required 10–12 routine transfusions annually. Consequently, this patient falls into the category of “severe,” according to the Mahidol severity score [6].
Since it is so close to New Delhi, India’s capital city, the western part of Uttar Pradesh has a special geographical location. As a result, this city is referred to as the national capital region (NCR), and it has attracted people of various races and ethnic groups from various Indian states. A national meta-analysis found that 52 mutations account for 97.5% of all β-thalassemia alleles, with IVS I-5 (G>C) being the most common (54.7%). IVS I-5 (G>C), IVS I-1 (G>T), 619-bp del, CD41/42 (-TCTT), and CD 8/9 (+G) accounted for 90% of all mutations among the 52 mutations [7, 8, 9]. We describe a case with an unusual combination of four distinct mutations in the β-globin gene (CD3 [T>C], CD41/42 [-CTTT], IVS II-16 [G>C], and IVS II-666 [C>T]) in our case study (Table 1, Fig. 1).
The CD3 (T>C), rs713040 is a clinically benign variant (Clinvar), in which the codon CAC is changed from CAT and amino acid histidine is substituted for amino acid histidine, resulting in no change in the protein sequence (Table 1). However, in India, this variant is one of the most uncommon, although it is reported in Odisha Province, and in the Bangladeshi population [10, 11]. The CD41/42 (-CTTT) mutation (rs80356821) in exon 2 of the β-globin gene is a four-nucleotide deletion that induces frameshift and stops the development of β-globin protein, and was first reported in an Asian Indian with β0-thalassemia [7]. Clinically pathogenic codon 41/42 (-CTTT) mutations lead to significantly higher HbA2 levels (> 4.0) and lower MCV and MCH levels [4]. Another two variants, IVS II-16 (G>C), and IVS II-666 (C>T), were found in the intron 2 region of the gene. Previously, IVS II-16 (G>C) and IVS II-666 (C>T) have been recognized by AvaII and SspI, respectively [12, 13]. Both the variants have been reported in many previous studies from East India, Bangladesh, Saudi Arabia, Egypt, and Palestine [11, 12, 14, 15, 16, 17, 18, 19, 20, 21]. However, these polymorphisms are not involved in the gene expression and are assumed to be benign. However, when these benign variants are found in association with HbS, they act as likely pathogenic variants and convert the HbS phenotype to transfusion-dependent Hb S/β+-thal [22, 23]. Another report from Turkey suggests that IVS II-16 (G>C), along with IVS II-74 (T>G), are “likely pathogenic” polymorphisms because patients with this polymorphism exhibit low levels of MCV and HbA [24]. Thus, it can be hypothesized that the IVS II-16 and IVS II-666 variants may be exerting a modulatory effect by possibly affecting mRNA splicing, depending on their proximity to exon-intron boundary, and should not be regarded as innocuous. However, more next generation/whole genome sequencing research may be helpful to overcome the ambiguity, and to evaluate the true clinical significance of these variants [25]. Additionally, inheritance of such benign variants may have significance in tracing the demographic migration of humans over time [26, 27].
In this report, the child patient was affected with β-thalassemia major, having low levels of MCH and MCV. HbA2 was > 3.0 % and CD41/42 (-CTTT) is the only apparent pathogenic variant that causes ineffective erythropoiesis and hemolytic anemia (Ithanet).
Conclusion
Direct gene sequencing helps in the detection of unknown and unusual gene mutations. Although all four variants (CD3 [T>C], CD41/42 [-CTTT], IVS II-16 [G>C], and IVS II-666 [C>T] have been identified in several studies, this is the first study to describe an uncommon combination (a rare compound heterozygous condition) of four distinct changes in a single gene. CD41/42 (-CTTT) is the only well-established pathogenic mutation that causes deleterious phenotypes (β0). The other three variants are considered as benign polymorphisms, but some reports suggest that IVS II-16 (G>C) is likely a pathogenic variant. A large number of mutations identified in the population cohorts were also included in the natural selection study. These sorts of variants (harmless and dangerous) in combination may play a significant role in population migration studies. As a result, the family histories of people who have unusual sets of variants may be useful in the study of human migration.
Availability of data and materials
Not applicable.
Abbreviations
- IVS:
-
Intervening sequence
- HBB:
-
Beta hemoglobin
- CD:
-
Codon
- BMI:
-
Body mass index
- HbF:
-
Fetal hemoglobin
- HbA:
-
Adult hemoglobin
- MCV:
-
Mean corpuscular volume
- MCH:
-
Mean corpuscular hemoglobin
- CBC:
-
Complete blood count
References
Cunningham MJ, Macklin EA, Neufeld EJ, Cohen AR, Thalassemia Clinical Research Network. Complications of beta-thalassemia major in North America”. Blood. 2004;104(1):34–9. https://doi.org/10.1182/blood-2003-09-3167.
Weatherall DJ, Clegg JB. Inherited haemoglobin disorders: an increasing global health problem. Bull World Health Organ. 2001;79(8):704–12.
Angastiniotis M, Modell B. Global epidemiology of hemoglobin disorders. Ann N Y Acad Sci. 1998;850(June):251–69. https://doi.org/10.1111/j.1749-6632.1998.tb10482.x.
Chauhan W, Afzal M, Zaka-Ur-Rab Z, Noorani MS. A novel frameshift mutation, deletion of HBB:C.199_202delAAAG [Codon 66/67 (-AAAG)] in β-thalassemia major patients from the western region of Uttar Pradesh, India. Appl Clin Genet. 2021;14:77–85. https://doi.org/10.2147/TACG.S294891.
George E, Li HJ, Fei YJ, Reese AL, Baysal E, Cepreganova B, Wilson JB, Gu LH, Nechtman JF, Stoming TA. Types of thalassemia among patients attending a large university clinic in Kuala Lumpur, Malaysia. Hemoglobin. 1992;16(1–2):51–66. https://doi.org/10.3109/03630269209005676.
Sripichai O, Makarasara W, Munkongdee T, Kumkhaek C, Nuchprayoon I, Chuansumrit A, Chuncharunee S, et al. A scoring system for the classification of β-thalassemia/Hb E disease severity. Am J Hematol. 2008;83(6):482–4. https://doi.org/10.1002/ajh.21130.
Kazazian HH, Orkin SH, Antonarakis SE, Sexton JP, Boehm CD, Goff SC, Waber PG. Molecular characterization of seven beta-thalassemia mutations in Asian Indians. EMBO J. 1984;3(3):593–6.
Sinha S, Black ML, Agarwal S, Colah R, Das R, Ryan K, Bellgard M, Bittles AH. Profiling β-thalassaemia mutations in India at state and regional levels: implications for genetic education, screening and counselling programmes. HUGO J. 2009;3(1–4):51–62. https://doi.org/10.1007/s11568-010-9132-3.
Thein SL, Hesketh C, Wallace RB, Weatherall DJ. The molecular basis of thalassaemia major and thalassaemia intermedia in asian indians: application to prenatal diagnosis. Br J Haematol. 1988;70(2):225–31. https://doi.org/10.1111/j.1365-2141.1988.tb02468.x.
Ayub I, Mustak MM, Moosa GS, Khan W, Khan H, Yeasmin S. Mutation analysis of the HBB gene in selected bangladeshi beta-thalassemic individuals: presence of rare mutations. Genet Test Mol Biomarkers. 2010;14(3):299–302. https://doi.org/10.1089/gtmb.2009.0160.
Sahoo SS, Biswal S, Dixit M. Distinctive mutation spectrum of the HBB gene in an urban Eastern Indian population. Hemoglobin. 2014;38(1):33–8. https://doi.org/10.3109/03630269.2013.837394.
Akhavan-Niaki H, Banihashemi A, Azizi M. Beta globin frameworks in thalassemia major patients from North Iran. Iran J Pediatr. 2012;22(3):297–302.
Hashemi-Soteh MB, Mousavi SS, Tafazoli A. Haplotypes inside the beta-globin gene: use as new biomarkers for beta-thalassemia prenatal diagnosis in North of Iran. J Biomed Sci. 2017;24(1):92. https://doi.org/10.1186/s12929-017-0396-y.
El-Shanshory MR, Hagag AA, Shebl SS, Badria IM, Abd Elhameed AH, Abd El-Bar ES, Al-Tonbary Y, et al. Spectrum of beta globin gene mutations in Egyptian children with β-Thalassemia. Mediterr J Hematol Infect Dis. 2014;6(1):e2014071. https://doi.org/10.4084/MJHID.2014.071.
Gazi N, Begum R, Akhter H, Shamim Z, Rahim M, Chaubey G. The complete spectrum of Beta (β) thalassemia mutations in Bangladeshi population. Austin Biomark Diagn 3 (April). 2016.
Kulkarni GD, Kulkarni SS, Kadakol GS, Kulkarni BB, Kyamangoudar PH, Lakkakula BVKS, et al. Molecular basis of β-Thalassemia in Karnataka, India. Genet Testing Mol Biomark. 2012;16(2):138–41. https://doi.org/10.1089/gtmb.2011.0035.
Sirdah MM, Sievertsen J, Al-Yazji MS, Tarazi IS, Al-Haddad RM, Horstmann RD, Timmann C. The spectrum of β-thalassemia mutations in Gaza Strip, Palestine. Blood Cells Mol Dis. 2013;50(4):247–51. https://doi.org/10.1016/j.bcmd.2012.12.004.
Hanafi S, Hassan R, Bahar R, Abdullah WZ, Johan MF, Rashid ND, et al. Multiplex amplification refractory mutation system (MARMS) for the detection of β-globin gene mutations among the transfusion-dependent β-thalassemia Malay patients in Kelantan, Northeast of Peninsular Malaysia. Am J Blood Res. 2014;4(1):33–40.
Borgio JF, AbdulAzeez S, Naserullah ZA, Al-Jarrash S, Al-Ali RA, Al-Madan MS, Al-Muhanna F, et al. Mutations in the β-globin gene from a Saudi population: an update. Int J Lab Hematol. 2016;38(2):e38–40. https://doi.org/10.1111/ijlh.12463.
Alaithan MA, AbdulAzeez S, Borgio JF. A comprehensive review of the prevalence of beta globin gene variations and the co-inheritance of related gene variants in Saudi Arabians with beta-thalassemia. Saudi Med J. 2018;39(4):329–35. https://doi.org/10.15537/smj.2018.4.21360.
Aldakeel SA, Ghanem NZ, Al-Amodi AM, Osman AK, Al Asoom LI, Ahmed NR, Almandil NB, Akhtar MS, Azeez SA, Borgio JF. Identification of seven novel variants in the β-globin gene in transfusion-dependent and normal patients. Arch Med Sci. 2019;16(2):453–59. https://doi.org/10.5114/aoms.2019.84825.
Uçucu S, Karabıyık T, Azik FM. IVS-II-16 (G>C) (HBB: C.315+16G>C) or IVS-II-666 (C>T) (HBB: C.316-185C>T) mutations trigger an Hb S (HBB: C.20A>T)/Β+-thalassemia phenotype in an Hb S trait patient. Hemoglobin. 2021;45(4):225–7. https://doi.org/10.1080/03630269.2021.1965620.
Vucak J, Turudic D, Milosevic D, Bilic M, Salek Z, Rincic M, Bilic E. Genotype-phenotype correlation of β-thalassemia in croatian patients: a specific HBB gene mutations. J Pediatr Hematol Oncol. 2018;40(2):e77-82. https://doi.org/10.1097/MPH.0000000000001039.
Hocaoglu-Emre FS, Yenmis G, Saribal D, Yakicier C. IVSII-74 T>G: as harmless as we thought? Turk J Biochem. 2019;44(1):41–6. https://doi.org/10.1515/tjb-2018-0332.
Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, Aston M, Esterling L, Manley S, Ross T. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 2018;320(12):1266–74. https://doi.org/10.1001/jama.2018.13152.
Choudhury A, Aron S, Botigué LR, Sengupta D, Botha G, Bensellak T, Wells G, et al. High-depth African genomes inform human migration and health. Nature. 2020;586(7831):741–8. https://doi.org/10.1038/s41586-020-2859-7.
Lindenau JD, Wagner SC, de Castro SM, Hutz MH. The effects of old and recent migration waves in the distribution of HBB*S globin gene haplotypes. Genet Mol Biol. 2016;39(4):515. https://doi.org/10.1590/1678-4685-GMB-2016-0032.
Acknowledgements
The authors are grateful to the Chairman, Department of Zoology, AMU, and also the Dean faculty of life science, Professor Mohammad Afzal and Head, for providing necessary laboratory facilities. The Indian Council of Medical Research (ICMR), India, is highly acknowledged for providing a fellowship to Waseem Chauhan during the study.
Funding
The author(s) received no financial support for the research, authorship and/or publication of this article.
Author information
Authors and Affiliations
Contributions
WC performed the analysis, conceptualization, data curation, experimental work, and writing the manuscript. RF performed the manuscript writing and editing and experimental work. MA performed the supervision, conceptualization, analysis, and final reviewing and editing. ZZR performed patient handling, data curation, and reviewing and editing the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethical approval was provided by the Institutional Ethical Committee (Aligarh Muslim University). Written informed consent was obtained from the guardian of the patient for participation in this study.
Consent for publication
Written informed consent was obtained from the legal guardians of the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Competing interests
In relation to the research, authorship, and/or publication of this article, the author(s) declared no potential competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chauhan, W., Fatma, R., Zaka-ur-Rab, Z. et al. Direct sequencing of β-globin gene reveals a rare combination of two exonic and two intronic variants in a β-thalassemia major patient: a case report. J Med Case Reports 16, 362 (2022). https://doi.org/10.1186/s13256-022-03605-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13256-022-03605-2