Abstract
Objective
This study aimed to extract radiomics features from MRI using machine learning (ML) algorithms and integrate them with clinical features to build response prediction models for patients with spinal metastases undergoing stereotactic body radiotherapy (SBRT).
Methods
Patients with spinal metastases who were treated using SBRT at our hospital between July 2018 and April 2023 were recruited. We assessed their response to treatment using the revised Response Evaluation Criteria in Solid Tumors (version 1.1). The lesions were categorized into progressive disease (PD) and non-PD groups. Radiomics features were extracted from T1-weighted image (T1WI), T2-weighted image (T2WI), and fat-suppression T2WI sequences. Feature selection involved intraclass correlation coefficients, minimal-redundancy-maximal-relevance, and least absolute shrinkage and selection operator methods. Thirteen ML algorithms were employed to construct the radiomics prediction models. Clinical, conventional imaging, and radiomics features were integrated to develop combined models. Model performance was evaluated using receiver operating characteristic (ROC) curve analysis, and the clinical value was assessed using decision curve analysis.
Results
We included 194 patients with 142 (73.2%) lesions in the non-PD group and 52 (26.8%) in the PD group. Each region of interest generated 2264 features. The clinical model exhibited a moderate predictive value (area under the ROC curve, AUC = 0.733), while the radiomics models demonstrated better performance (AUC = 0.745–0.825). The combined model achieved the best performance (AUC = 0.828).
Conclusion
The MRI-based radiomics models exhibited valuable predictive capability for treatment outcomes in patients with spinal metastases undergoing SBRT.
Critical relevance statement
Radiomics prediction models have the potential to contribute to clinical decision-making and improve the prognosis of patients with spinal metastases undergoing SBRT.
Key points
• Stereotactic body radiotherapy effectively delivers high doses of radiation to treat spinal metastases.
• Accurate prediction of treatment outcomes has crucial clinical significance.
• MRI-based radiomics models demonstrated good performance to predict treatment outcomes.
Graphical Abstract

Similar content being viewed by others
Introduction
Bone metastases commonly affect the spine [1]. The number of patients presenting with spinal metastases is rising because of increasing cancer incidence and life expectancy [2, 3]. Spinal metastases can lead to pain, vertebral compression fractures, and compression of the spinal cord or nerve roots, which significantly affect patients’ quality of life [4].
The advanced radiotherapy technology, stereotactic body radiotherapy (SBRT), delivers highly conformal and ablative doses to extracranial target lesions [5]. Compared with palliative radiotherapy, SBRT more accurately irradiates spine metastatic lesions using ablative doses, while reducing radiation-induced injury risk to the minimum [6]. The intense antitumor effects of ablative irradiation with SBRT result in a high tumor control rate [7]. Studies suggest that imaging-based local control rates range from 57 to 100% [7,8,9].
Prediction of outcomes in patients with spinal metastases treated with SBRT could assist in treatment decision-making and in managing prognostic expectations. For those patients who do not achieve local control, the treatment plan may be altered to include systemic therapy and surgery. Previous studies have found that some clinical features such as the primary tumor site, polymetastatic disease, performance status, and pain level are associated with prognosis after SBRT in patients with spinal metastases [10,11,12,13]. However, the relationship between imaging features and patient prognosis is unknown.
MRI is a commonly used imaging method before patients receive SBRT and might have the potential for prognostic prediction. MRI has a high soft-tissue resolution, allowing for better visualization of spinal metastases and optimization of the accuracy of target volume delineation for SBRT compared with CT [14]. With recent advances in computer-aided diagnosis, quantitative image methods together with machine learning (ML) algorithms, which can extract large-scale data from medical images, might provide valuable prognostic information [15].
Therefore, in this study, we aimed to apply ML methods to extract high-throughput radiomics features from MRI, selecting optimal radiomics features integrated with clinical features to build a response prediction model for SBRT-treated patients with spinal metastases.
Materials and methods
Patient selection
The ethics committee of our hospital approved this prospective study. We recruited consecutive patients with spinal metastases who received SBRT at our institution from July 2018 to April 2023. All patients provided informed consent.
The inclusion criteria comprised: (1) a diagnosis of spinal metastases based on pathological biopsy or imaging, and (2) undergoing MRI within 1 week before receiving SBRT. The exclusion criteria comprised: (1) the patient’s target lesion area had received previous radiotherapy or surgery, (2) the patient had received chemotherapy within 1 month, (3) the patient was unable or refused to cooperate with CT and MRI examinations, (4) the patient was lost to follow-up, or (5) the image quality was poor, making analysis impossible.
MRI data acquisition
A 3.0-T GE Discovery MR 750 (GE Healthcare) MRI scanner was used, which had a phased-array body coil with eight channels. The following MRI sequences were performed: sagittal T1-weighted image (T1WI) (repetition time (TR): 400–750 ms; echo delay time (TE): 8–25 ms), sagittal T2WI (TR: 2500–4000 ms; TE: 120–140 ms), and sagittal fat-suppression T2WI (FS-T2WI) (TR: 2500–4000 ms; TE: 100–120 ms), transverse T2WI (TR: 2500–4000 ms; TE: 100–120 ms).
SBRT procedures and treatment outcome assessment
The treatment platform comprised the CyberKnife stereotactic radiotherapy system (Accuray Inc). Before treatment planning CT, patients were immobilized using either the BodyFix system for thoracic, lumbar, and sacral spine lesions, or a customized thermoplastic mask for cervical spine lesions. According to the Consensus Guidelines of the International Spine Radiosurgery Consortium [16], target delineation was conducted on co-registered MRI and CT datasets. A physicist formulated the radiotherapy plan using the Multiplan System and an X-sight spine tracking system was used to track lesions. The determination of radiotherapy dosage was based on factors such as histopathology, tumor location, and the tolerance dose of adjacent organs.
Treatment outcome assessment was assessed according to the Revised Response Evaluation Criteria in Solid Tumors (RECIST; version 1.1) [17], which divided the lesions into four groups: complete response (CR), partial response (PR), progressive disease (PD), and stable disease (SD). To facilitate the analysis, we classified the lesions into the PD group and non-PD group (including CR, PR, and SD).
Clinical data and conventional imaging features collection
We collected clinical information comprising age, sex, location of the primary tumor (lung carcinoma /abdominal tumor/others), the number of spinal lesions (single/multiple), the presence of lymph node metastasis, the presence of visceral metastasis, the dose of radiation (30–40 Gy/5 fractions, 24–30 Gy/3 fractions, 18–24 Gy/1 fraction, or others), pain score (11-point numerical rating scale; 0–3/4–6/7–10) [18], Karnofsky performance status (KPS) score (80–100/0–70) [19], and spine instability neoplastic score (SINS; 0–6/7–12/13–18) [20].
The following conventional imaging features were collected: location (sacral/lumbar/thoracic/cervical), pattern of bone destruction (osteoblastic/osteolytic/ mixed), presence of a soft-tissue mass, involvement of the vertebral body, involvement of the pedicle, involvement of the lamina, presence of vertebral compression, and epidural spinal cord compression scale (Bilsky scale; 0–1/2–3) [21].
Tumor segmentation
Two radiologists with over 5 years of experience, who were blinded to treatment outcomes, manually performed the segmentation of the tumor. The Digital Imaging and Communications in Medicine (DICOM) images were imported into the uAI Research Portal platform 1.1 (United Imaging Intelligence, Co., Ltd.) to perform image segmentation.
The region of interest (ROI) should include as much of the visible gross tumor as possible. Initially, the ROI was manually delineated on sagittal FS-T2WI. Subsequently, the ROI was replicated on sagittal T1WI and sagittal T2WI sequences and adjusted manually to ensure accuracy by redefining any problematic ROI.
Image pre-processing and feature extraction
In the image pre-processing stage, we used B-spline interpolation resampling for image voxel size normalization, and all image sets were resampled to isotropic voxel size of 1 × 1 × 1 mm. Z-score intensity normalization was applied to reduce the impact of variability in image intensities on the stability of the radiomics features.
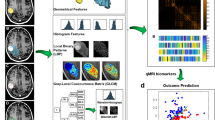
Feature extraction was performed using the uAI Research Portal platform 1.1 (United Imaging Intelligence, Co., Ltd.). Most features defined in this platform comply with feature definitions as described by the Image Biomarker Standardization Initiative (IBSI) [22].
The following classes of radiomics features were obtained from the original images: (1) First Order Statistics (19 features); (2) Shape-based (3D) (16 features); (3) Shape-based (2D) (10 features); (4) Gray Level Co-occurrence Matrix (GLCM, 24 features); (5) Gray Level Run Length Matrix (GLRLM, 16 features); (6) Gray Level Size Zone Matrix (GLSZM, 16 features); (7) Neighbouring Gray Tone Difference Matrix (NGTDM, 5 features); (8) Gray Level Dependence Matrix (GLDM, 14 features).
Furthermore, in order to increase the dimensionality of the dataset and improve the predictive performance of the models, we applied 17 different filters, including AdditiveGaussiannoise, Bilateral, BinomialBlurImage, BoxMean, BoxSigmaImage, CurvatureFlow, DiscreteGaussian, LaplacianSharpening, Mean, Median, Normalize, Recursive Gaussian, ShotNoise, SmoothingRecursiveGaussian, SpeckleNoise, LoG, and Wavelet to generate filtered images from the original ones. All classes of features, except for shape-based features, were computed on both the original and filtered images.
Feature selection and prediction model development
Feature selection includes several steps. First, to ensure inter-observer reliability for the features extracted from the ROIs drawn by two radiologists, intraclass correlation coefficients (ICCs) were analyzed. Features with an ICC > 0.75 were deemed reliable and could be selected for model construction. Next, the minimal-redundancy-maximal-relevance (mRMR) framework was used to identify the most relevant features to tumor classification and eliminate redundant features [23]. This produced the top 100 highly relevant and least redundant features. Finally, we used the least absolute shrinkage and selection operator (LASSO) regression model, incorporating 10-fold cross-validation, to select features with nonzero coefficients [24]. In addition to performing feature selection on each sequence (T1WI, T2WI, and FS-T2WI sequences), feature selection was also performed across all three combined sequences (ALL sequences).
The final selected radiomics features were applied to 13 ML algorithms including AdaBoost, XGBoost, bagging decision trees (Bagging), decision tree classifiers (DT), gaussian processes (GP), gradient boosted decision trees (GBDT), k-nearest neighbor (KNN), logistic regression (LR), partial least squares discriminant analysis (PLS-DA), quadratic discriminant analysis (QDA), random forest (RF), stochastic gradient descent (SGD), and support vector machine (SVM) to construct the radiomics prediction models.
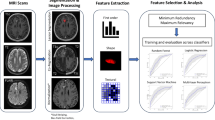
Next, we incorporated clinical, conventional imaging, and radiomics features into the ML algorithms to construct the combined model. The workflow of the predictive model construction is shown in Fig. 1.
The workflow of prediction model construction. a Tumor segmentation was performed on T1WI, T2WI, and FS-T2WI. b Quantitative features were extracted from each ROI. c Feature selection was conducted to reduce feature dimensionality and enhance prediction performance. d and e Three types of prediction models were constructed and evaluated
Statistical analysis
The mean ± standard deviation (SD) was used to describe variables with a normal distribution. Median values and ranges were used to describe variables with a non-normal distribution. Proportions described categorical variables. Upon analysis using univariate logistic regression, we selected clinical variables with p < 0.20, which were carried forward for analysis using multivariate logistic regression. Odds ratios (ORs) with 95% confidence intervals (CIs) were obtained for each clinical variable. The performances of each model were compared using the area under the curve (AUC) of the receiver operating characteristic (ROC) curve. Additionally, we carried out decision curve analysis (DCA) at various threshold probabilities to assess the net benefits of each model and to determine their clinical applicability. SPSS software version 26.0 (IBM Corp.) and R version 4.2.0 (R Foundation for Statistical Computing) were used to perform the statistical analyses. Statistical significance was considered at p-values < 0.05.
Results
Study cohort
We included 194 patients in the study, based on the inclusion and exclusion criteria (Fig. 2). The study population comprised 108 males and 86 females (mean age = 56.4 ± 15.3 years). There were 142 (73.2%) lesions in the non-PD group and 52 (26.8%) lesions in the PD group. Table 1 provides the detailed clinical and conventional imaging features for the entire cohort.
Clinical and conventional imaging feature-based model performance
Univariate and multivariate logistic regression analyses indicated that the number of spinal lesions, pain score, KPS score, and Bilsky grade were independent predictors of PD (see Table 2 for details). PD was more likely in patients with multiple spinal lesions (OR = 2.13, 95% CI: 1.01–4.52, p = 0.048), a high pain score (OR = 4.839, 95% CI: 1.40–16.79, p = 0.013), a low KPS score (OR = 2.17, 95% CI: 1.04–4.52, p = 0.038), and a high Bilsky grade (OR = 2.49, 95% CI: 1.17–5.28, p = 0.018). The clinical prediction model was constructed using these four features. The AUC for the clinical prediction model in predicting PD was 0.733 (Fig. 3).
Radiomics model and combined model performances
A total of 2264 features were generated for each ROI. According to the standard of ICC > 0.75, rates of stable features for T1WI, T2WI, and FS-T2WI sequences were 66.3%, 81.6%, and 79.0%, respectively.
Overall, the radiomics model established using features from all three MRI sequences outperformed those based on a single sequence. The optimal models constructed based on T1WI, T2WI, and FS-T2WI sequences achieved AUC values of 0.779 (QDA), 0.823 (GP), and 0.745 (QDA) respectively. The optimal model based on ALL sequences attained an AUC of 0.825 (GP, Fig. 3).
After incorporating clinical features, the performance of the combined model improved slightly. The best-performing model was based on ALL sequences using the GP algorithm (AUC = 0.828, Fig. 3). The diagnostic performance of the optimal ML classifier for each model is shown in Table 3. Decision curves revealed that the combined model exhibited the greatest net benefit in predicting PD (Fig. 4).
Decision curves for three models in the test set. Decision curve analysis demonstrates that the curves of the clinical, radiomics, and combined models all appear above the reference lines, indicating that these models provide a net benefit to improve clinical decision-making for patients. The radiomics and combined models exhibit higher net benefit compared with that of the clinical model
Discussion
Predicting treatment outcomes for spinal metastases undergoing SBRT is challenging; however, certain clinical and radiological features show potential predictive value. Our research identified the Bilsky grade, KPS score, pain score, and the number of spinal lesions as independent PD predictors.
The number of spinal lesions affected treatment outcome: patients with multiple spinal lesions had a higher risk of PD compared to those with a single lesion, which agreed with previously published results [10, 11]. As a local treatment, SBRT is particularly suitable for patients with a single spinal metastasis. Tree et al. [25] reviewed existing evidence and recommended that SBRT should be considered for patients with isolated metastases. We observed that compared with that of patients with lower pain scores, those with higher scores were more prone to PD, consistent with prior research [10]. We speculated that the reason for this phenomenon might be related to the stage of the tumor, because pain is typically not significant in the early stages of the tumor, while it becomes intense in the advanced stages. Therefore, patients with more severe pain are more likely to experience PD. In addition, pain-induced physical limitations, emotional instability, sleep disorders, and dietary issues might also affect patient prognosis. We observed a higher probability of PD in patients with lower KPS scores, consistent with previous research [12, 26]. The KPS score reflects the patient’s condition, with a lower score often indicating poorer overall health. This can be a sign of serious underlying health issues or symptoms. Consequently, patients with lower KPS scores might experience more difficulties and complications during SBRT, leading to a less favorable response to treatment and a worse prognosis. Based on T2WI, the Bilsky grade assesses the severity of spinal stenosis. Patients with Bilsky grades 2–3, indicating severe spinal stenosis, tended to experience PD more frequently than those with Bilsky grades 0–1 (mild stenosis), consistent with a previous study [11]. This might have been caused by the presence of larger tumors and their proximity to the spinal cord, which would limit the radiation dose. Consequently, SBRT might not be suitable for patients with severe spinal stenosis, particularly those with Bilsky grade 3 and significant neurological symptoms. Surgical treatment should be considered in such cases [27].
The clinical prediction model built based on the selected features showed a decent predictive value (AUC = 0.733). In comparison, the radiomics models constructed using MRI demonstrate better performance (AUC = 0.745–0.825). Radiomics techniques can extract a vast amount of information, providing a more comprehensive and detailed description of lesion characteristics [28]. By capturing subtle changes within the lesion, radiomics features can provide a more precise depiction of the complex physiopathological mechanisms of tumors. Therefore, the radiomics models based on MRI have significant value in predicting the treatment outcome of SBRT. Furthermore, when clinical features were incorporated, the combined models contained more valid information to predict PD, leading to a further improvement in performance (AUC = 0.828).
ML algorithms play a vital role as indispensable tools in radiomics. By extensively learning and training on a vast dataset, ML algorithms can reveal the association between image biomarkers and treatment outcomes [29]. When choosing the most suitable ML algorithm, it is vital to account for factors including data characteristics, task type, data scale, algorithm efficiency, and predictive performance [30]. Therefore, the optimal choice depends on specific circumstances. In our study, we utilized 13 ML algorithms to construct the predictive models. We found that the performances of the GP and QDA algorithms were better than those of other ML algorithms. We speculated that both GP and QDA share common characteristics of being able to handle nonlinear relationships between features and are applicable to small sample sizes without requiring large amounts of training data [31, 32].
As far as we know, this was the first study to employ MRI-based radiomics models to predict the treatment outcome post-SBRT in patients with spinal metastases. Previous studies have mainly investigated the association between treatment outcomes and clinical features [11, 12, 33]. Additionally, one study employed CT-based radiomics to predict patient’s pain response [34].
There were limitations associated with the present study. Firstly, this was a single-center study involving a limited number of cases. Sample size can affect the performance and generalization ability of ML algorithms. Therefore, large-scale multicenter studies should be carried out to gain more corroborative evidence for clinical applications. Secondly, we could only construct models based on MRI features because the majority of the patients did not receive CT scans before treatment. This was because MRI provides better visualization of spinal metastases due to its high soft-tissue resolution and optimal target volume delineation for SBRT compared with CT. From an economic aspect, except for those initially identified with metastases on CT, only pretreatment MRI was performed in our cohort. The value of radiomics models based on CT, as well as other imaging examinations, such as PET and functional MRI, deserves further exploration.
Conclusion
In conclusion, MRI is commonly utilized as an imaging modality before SBRT, enabling a comprehensive evaluation of target lesions. Through the analysis of radiomic features in MRI, our constructed models could predict treatment outcomes following SBRT in spinal metastases. Furthermore, the incorporation of clinical features further improved the performance of the models. These predictive models should aid clinicians’ decision-making and will contribute to improved prognosis of patients suffering from spinal metastases.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Bagging:
-
Bagging decision trees
- CI:
-
Confidence interval
- DT:
-
Decision tree classifiers
- FS-T2WI:
-
Fat-suppression T2-weighted image
- GBDT:
-
Gradient-boosted decision trees
- GP:
-
Gaussian processes
- ICC:
-
Intraclass correlation coefficient
- KNN:
-
k-nearest neighbor
- KPS:
-
Karnofsky performance status
- LR:
-
Logistic regression
- ML:
-
Machine learning
- OR:
-
Odds ratio
- PD:
-
Progressive disease
- PLS-DA:
-
Partial least squares discriminant analysis
- QDA:
-
Quadratic discriminant analysis
- RF:
-
Random forest
- SBRT:
-
Stereotactic body radiotherapy
- SGD:
-
Stochastic gradient descent
- SVM:
-
Support vector machine
- T1WI:
-
T1-weighted image
- T2WI:
-
T2-weighted image
References
Coleman RE, Croucher PI, Padhani AR et al (2020) Bone metastases. Nat Rev Dis Primers 6:83
Allemani C, Matsuda T, Di Carlo V et al (2018) Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 391:1023–1075
Glicksman RM, Tjong MC, Neves-Junior WFP et al (2020) Stereotactic ablative radiotherapy for the management of spinal metastases: a review. JAMA Oncol 6:567–577
Thibault I, Chang EL, Sheehan J et al (2015) Response assessment after stereotactic body radiotherapy for spinal metastasis: a report from the SPIne response assessment in Neuro-Oncology (SPINO) group. Lancet Oncol 16:e595–603
Kollar L, Rengan R (2014) Stereotactic body radiotherapy. Semin Oncol 41:776–789
Kowalchuk RO, Waters MR, Richardson KM et al (2020) Stereotactic radiosurgery for the treatment of bulky spine metastases. J Neurooncol 148:381–388
Husain ZA, Sahgal A, De Salles A et al (2017) Stereotactic body radiotherapy for de novo spinal metastases: systematic review. J Neurosurg Spine 27:295–302
Sahgal A, Larson DA, Chang EL (2008) Stereotactic body radiosurgery for spinal metastases: a critical review. Int J Radiat Oncol Biol Phys 71:652–665
Sahgal A, Bilsky M, Chang EL et al (2011) Stereotactic body radiotherapy for spinal metastases: current status, with a focus on its application in the postoperative patient. J Neurosurg Spine 14:151–166
Zeng KL, Sahgal A, Tseng CL et al (2021) Prognostic factors associated with surviving less than 3 months vs greater than 3 years specific to spine stereotactic body radiotherapy and late adverse events. Neurosurgery 88:971–979
Zeng KL, Sahgal A, Husain ZA et al (2021) Local control and patterns of failure for “Radioresistant” spinal metastases following stereotactic body radiotherapy compared to a “Radiosensitive” reference. J Neurooncol 152:173–182
Park HJ, Kim HJ, Won JH, Lee SC, Chang AR (2015) Stereotactic body radiotherapy (SBRT) for spinal metastases: who will benefit the most from SBRT? Technol Cancer Res Treat 14:159–167
Chao ST, Koyfman SA, Woody N et al (2012) Recursive partitioning analysis index is predictive for overall survival in patients undergoing spine stereotactic body radiation therapy for spinal metastases. Int J Radiat Oncol Biol Phys 82:1738–1743
Prins FM, van der Velden JM, Gerlich AS et al (2017) Superior target delineation for stereotactic body radiotherapy of bone metastases from renal cell carcinoma on MRI compared to CT. Ann Palliat Med 6:S147–s154
Avanzo M, Wei L, Stancanello J et al (2020) Machine and deep learning methods for radiomics. Med Phys 47:e185–e202
Cox BW, Spratt DE, Lovelock M et al (2012) International Spine Radiosurgery Consortium consensus guidelines for target volume definition in spinal stereotactic radiosurgery. Int J Radiat Oncol Biol Phys 83:e597–605
Eisenhauer EA, Therasse P, Bogaerts J et al (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45:228–247
Karcioglu O, Topacoglu H, Dikme O, Dikme O (2018) A systematic review of the pain scales in adults: which to use? Am J Emerg Med 36:707–714
Puvanesarajah V, Lo SL, Aygun N et al (2015) Prognostic factors associated with pain palliation after spine stereotactic body radiation therapy. J Neurosurg Spine 23:620–629
Fisher CG, DiPaola CP, Ryken TC et al (2010) A novel classification system for spinal instability in neoplastic disease: an evidence-based approach and expert consensus from the Spine Oncology Study Group. Spine (Phila Pa 1976) 35:E1221–1229
Bilsky MH, Laufer I, Fourney DR et al (2010) Reliability analysis of the epidural spinal cord compression scale. J Neurosurg Spine 13:324–328
Zwanenburg A, Leger S, Vallières M, Lck SJR, Oncology (2016) Image biomarker standardisation initiative
Peng H, Long F, Ding C (2005) Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27:1226–1238
Tibshirani R (1996) Regression Shrinkage and Selection Via the Lasso. J R Stat Soc B (Methodological) 58:267–288. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x
Tree AC, Khoo VS, Eeles RA et al (2013) Stereotactic body radiotherapy for oligometastases. Lancet Oncol 14:e28–37
Luksanapruksa P, Buchowski JM, Hotchkiss W, Tongsai S, Wilartratsami S, Chotivichit A (2017) Prognostic factors in patients with spinal metastasis: a systematic review and meta-analysis. Spine J 17:689–708
Jabbari S, Gerszten PC, Ruschin M, Larson DA, Lo SS, Sahgal A (2016) Stereotactic body radiotherapy for spinal metastases: Practice Guidelines, Outcomes, and Risks. Cancer J 22:280–289
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Mayerhoefer ME, Materka A, Langs G et al (2020) Introduction to Radiomics. J Nucl Med 61:488–495
Uribe CF, Mathotaarachchi S, Gaudet V et al (2019) Machine learning in nuclear medicine: Part 1-Introduction. J Nucl Med 60:451–458
Rasmussen CE, Williams C (2006) Gaussian Process for Machine Learning
Solberg HE (1978) Discriminant analysis. CRC Crit Rev Clin Lab Sci 9:209–242
Yamada Y, Katsoulakis E, Laufer I et al (2017) The impact of histology and delivered dose on local control of spinal metastases treated with stereotactic radiosurgery. Neurosurg Focus 42:E6
Wakabayashi K, Koide Y, Aoyama T et al (2021) A predictive model for pain response following radiotherapy for treatment of spinal metastases. Sci Rep 11:12908
Funding
This study has received funding by National Natural Science Foundation of China (No 81971578, No81701648) and Key Clinical Projects of the Peking University Third Hospital (BYSY2018007).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by YC, SQ, and WZ. The first draft of the manuscript was written by YC, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Approval was granted by the Medical Science Research Ethics Committee of Peking University Third Hospital. Written informed consent was obtained from all subjects (patients) in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, Y., Qin, S., Zhao, W. et al. MRI feature-based radiomics models to predict treatment outcome after stereotactic body radiotherapy for spinal metastases. Insights Imaging 14, 169 (2023). https://doi.org/10.1186/s13244-023-01523-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13244-023-01523-5