Abstract
Background
Autism spectrum disorder (ASD) is still diagnosed through behavioral observation, due to a lack of laboratory biomarkers, which could greatly aid clinicians in providing earlier and more reliable diagnoses. Metabolomics on human biofluids provides a sensitive tool to identify metabolite profiles potentially usable as biomarkers for ASD. Initial metabolomic studies, analyzing urines and plasma of ASD and control individuals, suggested that autistic patients may share some metabolic abnormalities, despite several inconsistencies stemming from differences in technology, ethnicity, age range, and definition of “control” status.
Methods
ASD-specific urinary metabolomic patterns were explored at an early age in 30 ASD children and 30 matched controls (age range 2–7, M:F = 22:8) using hydrophilic interaction chromatography (HILIC)-UHPLC and mass spectrometry, a highly sensitive, accurate, and unbiased approach. Metabolites were then subjected to multivariate statistical analysis and grouped by metabolic pathway.
Results
Urinary metabolites displaying the largest differences between young ASD and control children belonged to the tryptophan and purine metabolic pathways. Also, vitamin B6, riboflavin, phenylalanine-tyrosine-tryptophan biosynthesis, pantothenate and CoA, and pyrimidine metabolism differed significantly. ASD children preferentially transform tryptophan into xanthurenic acid and quinolinic acid (two catabolites of the kynurenine pathway), at the expense of kynurenic acid and especially of melatonin. Also, the gut microbiome contributes to altered tryptophan metabolism, yielding increased levels of indolyl 3-acetic acid and indolyl lactate.
Conclusions
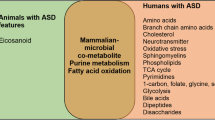
The metabolic pathways most distinctive of young Italian autistic children largely overlap with those found in rodent models of ASD following maternal immune activation or genetic manipulations. These results are consistent with the proposal of a purine-driven cell danger response, accompanied by overproduction of epileptogenic and excitotoxic quinolinic acid, large reductions in melatonin synthesis, and gut dysbiosis. These metabolic abnormalities could underlie several comorbidities frequently associated to ASD, such as seizures, sleep disorders, and gastrointestinal symptoms, and could contribute to autism severity. Their diagnostic sensitivity, disease-specificity, and interethnic variability will merit further investigation.
Similar content being viewed by others
Background
Autism spectrum disorder (ASD) represents a highly heterogeneous collection of neurodevelopmental conditions characterized by social and communication deficits, stereotypic and rigid patterns of behavior, restricted interests, and unusual sensory processing with onset in early childhood [1]. The prevalence of autism has increased significantly during the last two decades from 2–5/10,000 to 1:68 children [2, 3]. Changes in diagnostic criteria and increased attention by the medical community have certainly contributed to this trend [4]. Also, increasing parental age at conception has been shown to confer ASD risk [5], as well as some environmental factors, active especially during critical periods in prenatal/early postnatal neurodevelopment [6]. Finally, genetic susceptibility plays a prominent role in ASD pathogenesis through complex and heterogeneous underpinnings, ranging from rare variants endowed with full penetrance to common variants each explaining very small proportions of the overall phenotypic variance, either alone or through gene × environment interactions [7, 8].
Despite major advances in our understanding of the pathophysiology of ASD, this level of complexity and interindividual heterogeneity has largely hampered the translation of scientific knowledge into more effective clinical practices. ASD is still diagnosed exclusively through observation, standardized behavioral scales, and parental interviews; developmental trajectories of ASD children are periodically monitored but cannot be reliably predicted especially at an early age. Sensitive and specific quantitative biomarkers, measurable through laboratory, brain imaging, and/or electrophysiological techniques, could greatly aid clinicians in providing earlier diagnoses, more timely referrals to behavioral intervention programs, and evidence-based prognostic predictions [9].
Metabolomic technologies offer a sensitive means to search human biofluids for metabolite profiles potentially usable as biomarkers for neurodevelopmental disorders. A few studies have recently begun exploring the potential of urinary metabolomics in identifying ASD-specific metabolic patterns or in stratifying ASD patients into pathophysiologically meaningful subgroups [10–17]. Most studies have been performed on urines [10–16]; one study has explored blood plasma [17]. The analytical platforms most commonly used to identify and quantify metabolites are gas or liquid chromatography combined with mass spectroscopy (gas chromatography (GC)-mass spectroscopy (MS) and liquid chromatography (LC)-MS, respectively) [12, 16] and nuclear magnetic resonance spectroscopy (NMR) [10, 13, 14, 16, 18, 19]. In general, NMR displays greater speed and good reproducibility but also lower sensitivity compared to MS. Hence, MS- and NMR-based techniques should be viewed as complementary, not as superimposable approaches. An initial study, using 1H-NMR methods, showed an abnormal composition of urinary solutes indicative of perturbations in (a) the tryptophan/nicotinic acid metabolic pathway, (b) sulfur and amino acid metabolisms, and (c) gut microbiome, with an excess of several gut-derived co-metabolites [10]. Two other studies presumably assessing the same clinical sample with two different NMR-based technologies largely replicated these initial findings [13, 14]. Other studies using GC-MS, either alone [12, 15] or in combination with liquid chromatography [11], also identified perturbations in amino acid metabolism and gut microbial co-metabolites, as well as metabolic signatures of oxidative stress. Only one very recent study used both NMR and LC-MS, providing support for abnormalities in tryptophan metabolism, gut bacterial-derived compounds, purine and pyrimidine metabolism [16]. The only study exploring blood plasma reported metabolomic patterns compatible with (a) mitochondrial dysfunction, yielding reduced energy production and unbalanced redox status, (b) excess gut microbial co-metabolites, and (c) unbalances in various metabolic pathways, such as the Krebs cycle [17]. Collectively, metabolomic studies performed to this date suggest that autistic patients may share several metabolic abnormalities, especially involving some amino acid metabolisms, energy production, and oxidative stress, as well as the gut microbiome.
Moving from broad metabolic pathways to single compounds unveils inconsistencies between studies, which may stem from several potential confounds. Interethnic differences in the gut microbiota, stemming from differences in the nutrient composition of local diets, as well as age-related changes in both gut microbiota and human metabolism indeed require that case and control samples be tightly matched for these two variables. Age-related changes may be especially relevant to studies of ASD, where we have recently reported levels of urinary p-cresol to be elevated in autistic children compared to age-matched controls both in Italy and in France, but exclusively up until 8 years of age [20, 21]. Similar age-related changes in ASD have been previously described for other parameters, such as brain serotonin synthesis capacity [22, 23] and excessive head growth rates [24]. Finally, some studies have contrasted ASD patients with unrelated population controls [11, 14, 16, 17], while others have enrolled unaffected siblings as controls [15] and one study has used both [10]. These strategies are not equivalent, as first-degree relatives often fall within the broad autism spectrum (i.e., they display behavioral phenotypes intermediate between patients and population controls) [25]. In addition, siblings may carry protective gene variants with peculiar functional correlates, possibly distinct from the metabolic patterns of unrelated typically developing children.
Taking into consideration these methodological issues, in order to maximize the probability of reliably detecting differences in urinary metabolic patterns, we focused on autistic and unrelated typically developing children 2–8 years old, tightly matched by age, sex, Italian ancestry, and city of origin within the country [20]. To ensure broad metabolite detection coverage on urine samples, which comprise molecules generated both by human cells and by the gut microbiome, we employed hydrophilic interaction chromatography (HILIC)-LC-electrospray ionization (ESI)-MS, a technology particularly suitable to separate simple and complex mixtures of carbohydrates, amino acids, glycosides, and other natural polar products in biological fluids, such as human urine and plasma [26, 27]. Applying this experimental approach, urinary metabolites most significantly distinguishing autistic from typically developing children were found to primarily fall into the tryptophan and purine metabolic pathways.
Methods
Subjects
Thirty children with idiopathic ASD and thirty typically developing controls were recruited in Central and Northern Italy. These represent the vast majority of the 64 cases and controls aged 3–7 years assessed for urinary p-cresol in our previous study [20]. Their demographic and clinical characteristics are summarized in Additional file 1: Table S1. Diagnostic assessments and medical screening have been previously described [20] (also see Additional file 2 with Supplementary Methods). Tight sex- and age-matching (±1 year) was applied to recruit typically developing children devoid of any overt ASD symptomatology among the offspring of clinical/academic personnel [20]. Mean age (±SEM) of cases and controls was 4.83 ± 0.30 and 5.03 ± 0.32 years, respectively (Student’s t = −0.459, 58 df, P = 0.648, n.s.), and the M:F ratio was 22:8. All cases and controls were of Italian descent and matched by geographical area or city of origin.
Urine collection and metabolite extraction
First-morning urines were collected at home by parents using sterile containers untreated with preservatives and were brought to each clinical center the same morning in wet ice. Urine samples were then frozen, shipped in dry ice, and stored at −80 °C continuously until analysis.
Urinary specific gravity was measured by refractometry following centrifugation at 13,000g for 10 min) using a digital refractometer (Euromex Clinical Digital Refractometer RD.5712, NL) previously calibrated with LC-MS grade water.
Urine aliquots (200 μl) were mixed with 200 μl of methanol:acetonitrile:water (50:30:20), vortexed for 30 min at max speed at 4 °C and then centrifuged at 16,000g for 15 min at 4 °C. Supernatants were collected for metabolomic analysis. Quality controls (QCs) were obtained from a pooled mixture of 10 μl aliquots of all urine samples and were analyzed every 15 samples.
HILIC-UHPLC
Metabolite separation was performed as previously described [28], by hydrophilic interaction chromatography (HILIC) using the Ultimate 3000 Rapid Resolution HPLC system (Dionex, Sunnyvale, CA), featuring a binary pump and vacuum degasser, well-plate autosampler with a six-port micro-switching valve, and a thermostated column compartment. A Phenomenex Luna 3 μm HILIC 200 A (150 × 2.0 mm) column, protected by a HILIC 4 × 2.0 mm ID guard column (Phenomenex, Torrance, CA), was used to perform metabolite separation over a phase B-to-phase A gradient lasting 35 min. For the HILIC separation, mobile phase “A” consisted in 50 mM ammonium acetate mixed with acetonitrile (95:5, v/v), while eluent “B” was composed of a mixture of 50 mM ammonium acetate:water plus acetonitrile (95:5, v/v). Acetonitrile, formic acid, and HPLC-grade water were purchased from Sigma-Aldrich (St. Louis, MO).
Mass spectrometry
MS analysis was carried out on an electrospray hybrid quadrupole time-of-flight instrument MicroTOF-Q (Bruker-Daltonik, Bremen, Germany) equipped with an ESI ion source, as previously described [29]. Mass spectra for metabolite-extracted samples were acquired both in positive and in negative ion modes; only data produced in negative mode are shown, because more powerful in analyzing urinary samples. ESI capillary voltage was set at 4500 V (−) ion mode. The liquid nebulizer was set at 27 psi, and the nitrogen drying gas was set to a flow rate of 6 L/min. Dry gas temperature was maintained at 200 °C. Data were stored in centroid mode and acquired with a stored mass range of 50–1200 m/z. Instrument calibration was performed externally every day with 10 mM sodium hydroxide in 50% isopropanol: water, 0.1% formic acid. Automated internal mass scale calibration was performed through direct automated injection of the calibration solution at the beginning and at the end of each run by a six-port divert valve.
Data elaboration and statistical analysis
Data were normalized by urinary specific gravity, because creatinine excretion may be abnormally reduced in ASD children [30]. Replicates were exported as mzXML files and processed through MAVEN.52 (available at http://genomics-pubs.princeton.edu/mzroll/index.php?show=index) [31]. Mass spectrometry chromatograms were elaborated for peak alignment, matching and comparison of parent and fragment ions, and tentative metabolite identification (within a 10-ppm mass deviation range between observed and expected results against the imported Kyoto Encyclopedia of Genes and Genomes (KEGG) database). Representative examples of mass determination and MS/MS fragmentation graphs are presented for kynurenine, melatonin, and tryptophan in Additional file 3: Figure S1. Multivariate statistical analyses were performed on the entire metabolomics data set using the MetaboAnalyst 3.0 software (http://www.metaboanalyst.ca) [32], which also overviewed data variance structure in an unsupervised manner and produced scatter plots.
Orthogonal partial least squares discriminant analysis (OPLS-DA), which defines a predictive model that describes the direction of the maximum covariance between a dataset (X) and class membership (Y), was then used to maximize the difference in metabolic profiles between cases and controls [33, 34]. OPLS-DA was performed using the Excel add-in Multibase package (Numerical Dynamics, Japan; http://www.numericaldynamics.com/) by applying orthogonal signal correction on the metabolite concentrations shifted, log10 transformed, centered, and scaled to unit variance.
Performance of the optimal model was tested by a receiver operating characteristic (ROC) curve analysis and the validation data set, as performed using MetaboAnalyst 3.0 software (http://www.metaboanalyst.ca) [32].
For case-control contrasts of single urinary metabolites, significance threshold was held at a nominal P < 0.05 with no correction for multiple testing, because (a) differences in single metabolite concentrations were tested only following significant differences in pathway enrichment were detected, (b) intra-pathway variability of single metabolites is non-independent, and (c) also different metabolic pathways are not fully independent, as some metabolites fall into more than one pathway. Detailed and summary statistics are provided in Additional files 4 and 5.
Results
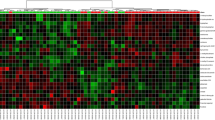
The urinary metabolomes of young autistic and typically developing children are largely distinguishable on the three-dimensional OPLS-DA plot depicting the first three principal components (PC), which together explain 31.4% of the total variance (Fig. 1; accuracy, Q2 and R2 data are shown in Additional file 6). Approximately 10,000 peaks per sample were obtained referring to the KEGG database; among them, 202 metabolites were analyzed more precisely and identified. The top 25 most discriminating metabolites between cases and controls were further defined based on “variable influence on the projection” (VIP) scores >1 (Fig. 2). ROC analysis using this set of 25 metabolites yielded an AUC = 0.893 (95% CI 0.72–0.96), as shown in Additional file 7. The “metabolome overview” obtained through metabolic pathway analysis (MetPA) shows tryptophan metabolism, purine metabolism, vitamin B6 metabolism, and phenylalanine-tyrosine-tryptophan biosynthesis as the four most perturbed metabolic pathways in ASD (Fig. 3).
Metabolic pathway analysis plot. Color intensity (white to red) reflects increasing statistical significance, while circle diameter covaries with pathway impact. The graph was obtained plotting on the y-axis the −log of p values from the pathway enrichment analysis and on the x-axis the pathway impact values derived from the pathway topology analysis
Given the relevance of tryptophan-derived compounds in many neural functions, tryptophan metabolism was assessed in greater detail at the level of specific intermediates (Fig. 4):
-
The kynurenine pathway displays increases in xanthurenic acid and especially in quinolinic acid, paralleled by a considerable decrease in kynurenic acid (Fig. 4, path A).
-
The serotonin pathway shows a significant decrease in melatonin and its catabolite N-acetyl-5-methoxytryptamine, which have the same molecular weight and thus fall under the same MS peak (Fig. 4, path B).
-
Bacterial degradation of tryptophan yields in ASD, compared to controls, prominently larger urinary concentrations of indoxyl sulfate and other indole derivatives, including indolyl-3-acetic acid and especially indolyl lactate (Fig. 4, paths C and D).
Also, purine metabolism was found to convey sizable discriminative power, because ASD cases display higher urinary concentrations of many purine metabolites compared to controls, including, among others, inosine, hypoxanthine, and xanthosine (Fig. 5).
Discussion
The present study reports significant urinary metabolomic differences between young children with idiopathic ASD and typically developing controls. At least some of the metabolic perturbations described here may reflect pathophysiologically meaningful abnormalities, possibly bearing functional consequences at the clinical level. Three strengths of the experimental design may have contributed to this positive outcome: (a) a focus on early infancy, by recruiting children within a relatively narrow age window precisely defined on the basis of previous urinary metabolic data [20, 21]; (b) the use of UHPLC-MS paired with HILIC, a very sensitive and reliable method ensuring maximum accuracy in the separation of small urinary solutes [26, 27]; (b) a pathway-centered approach, moving beyond the identification of single urinary ASD markers [10–17], as beautifully exemplified by urinary metabolomic studies of rodent models of ASD [35–37]. In particular, our recruitment strategy substantially differs from previous case-control study designs, minimizing age-dependent heterogeneity by setting data-driven age thresholds (i.e., 2–8 years old) [20, 21], and applying tight age and sex matching between cases and controls. This strategy seemingly circumvents sample size limitations which would apply to an unfocused and unmatched case-control design. Future replications obtained applying similar recruitment criteria will enhance confidence in the pathophysiological relevance and the interethnic generalizability of our findings.
The tryptophan metabolic pathway collectively displays the largest perturbations in ASD (Fig. 3). Over 90–95% of dietary l-tryptophan is usually metabolized along the kynurenine pathway, 1–2% is converted to serotonin, and approximately 4–6% undergoes bacterial degradation prior to gut absorption through the Na+-amino acid co-transporter B0AT1 (Slc6a19) [38, 39]. The latter pathway yields indole derivatives not produced by mammalian metabolism, such as indoxyl sulfate [40]. Hence, changes in urinary amounts of multiple metabolites provide more reliable evidence of perturbed tryptophan metabolism, as compared to determinations of single metabolites or tryptophan itself, which also suffer from reduced statistical power due to control for multiple testing (Figs. 4 and 5). In the urines of young autistic children, we have indeed observed a substantial increase of xanthurenic acid and especially of quinolinic acid, paralleled by a decrease in kynurenine and kynurenic acid (Fig. 4, path A). This pattern is extremely interesting but must be interpreted with some caution in the absence of parallel assessments of the cerebrospinal fluid (CSF). On the one hand, the enzymes responsible for the synthesis of quinolinic acid and xanthurenic acid are primarily expressed in the microglia and in macrophages, whereas the path leading to kynurenic acid is functional in astrocytes [41]. Hence, it would be tempting to speculate that these opposite trends between cases and controls reflect an abnormal activation of microglia, which has been repeatedly seen in ASD postmortem brains [42–44], even as early as at 4 years of age [45]. On the other hand, urinary levels of quinolinic acid and kynurenic acid reflect peripheral production of these compounds, which do not pass the blood-brain barrier [41]. However, 3-hydroxykynurenine does pass the blood-brain barrier [41]. Interestingly, urinary concentrations of metabolite downstream of this compound (quinolinic acid and xanthurenic acid) are elevated in autistic children, whereas metabolite upstream of 3-hydroxykinurenine (kynurenine and kynurenic acid) are higher among controls (Fig. 4, path A). Conceivably, these trends could reflect an outflow of 3-hydroxykynurenine from the central nervous system (CNS) into the systemic circulation, where macrophage activation presumably at the level of the gut or in other peripheral organs, can transform this compound into quinolinic acid and xanthurenic acid, as well as into nicotinic acid (NAD), in agreement with previous data [10]. It will thus be important to verify this metabolomic scenario in the CSF, because it could have at least two important clinical implications: (a) quinolinic acid acts as gliotoxin, proinflammatory mediator, and pro-oxidant molecule, boosting oxidative stress by stimulating microglia to release large amounts of NO and superoxide; (b) quinolinic acid exerts excitotoxic effects by acting as an N-methyl-d-aspartate (NMDA) receptor agonist, stimulating glutamate release, blocking glutamate reuptake into astrocytes, and reducing the activity of glutamine synthase; instead, kynurenic acid exerts neuroprotection via NMDA antagonism at the glycine binding site, as well as antioxidant effects [41, 46, 47]. In summary, the urinary metabolic imbalance documented here, if present also in the CNS, could favor enhanced oxidative stress and the well-known excitation>inhibition imbalance present in ASD, fostering seizures in as many as 20% of autistic individuals [48].
Another consequence of the preferential metabolization of tryptophan along the main branch of the kynurenine pathway is the relative decrease in the production of serotonin and melatonin (Fig. 4, path B). The serotonin pathway sees tryptophan being converted into 5-hydroxytryptophan (5-HTP) by tryptophan hydroxylase and onwards to 5-hydroxytryptamine (5-HT) or serotonin by 5-HTP decarboxylase. Serotonin can then be catabolized to 5-hydroxyindoleacetic acid (5-HIAA) or transformed into N-acetylserotonin by arylalkylamine N-acetyltransferase (AANAT). N-acetylserotonin is further methylated by N-acetylserotonin O-methyltransferase (ASMT) to generate the neurohormone 5-methyl-5-methoxy-N-tryptamine or melatonin. Decreases in the serotonin metabolite 5-HIAA are only modest, while urinary melatonin and its catabolite N-acetyl-5-methoxytryptamine display a more pronounced mean reduction (both share the same molecular weight and fall under the same MS peak, labeled in Fig. 4, path B, as “melatonin” only). This confirms previous assessments performed in plasma or urine [10, 49–52], while lending further support to blunted melatonin synthesis possibly due to reduced ASMT enzyme activity in ASD [53, 54]. Melatonin is synthesized and released by the pineal gland into the systemic circulation and readily passes the blood-brain barrier [55]. Its well-known role in circadian rhythmicity makes it an ideal candidate to explain the frequent occurrence, especially at the onset of ASD and during early infancy, of sleep disorders highly responsive to melatonin as a pharmacological therapy [56].
Metabolites produced by gut bacteria are well-represented also in our ASD sample, as in previous studies [10–17]. In addition to urinary p-cresol, found elevated in these same urine samples both here (Fig. 3) and previously using a different technology [20], we also detect a significant increase in indole derivatives of bacterial tryptophan including indolyl 3-acetic acid, indoxyl sulfate, and most prominently, indolyl lactate (Fig. 4, path C). Bacterial species expressing tryptophanase, the enzyme responsible for transforming tryptophan into indole derivatives, include Escherichia coli, Proteus vulgaris, Paracolobactrum coliform, Achromobacter liquefaciens, and Bacteroides spp. [40]. Once produced in the gut lumen, indole is absorbed, oxidized to indoxyl, conjugated with sulfate, and excreted as urinary indoxyl sulfate. About 3% of tryptophan entered with the diet is excreted as indoxyl sulfate [37]. Additional small amounts of tryptophan are converted into other indole derivatives found elevated here in ASD children, such as indolyl-3-acetic acid and indolyl lactate (Fig. 4, path C). The latter compound and indolyl 3-acetic acid are direct precursors of indolylacrylol glycine, found elevated in ASD by some [57] but not all studies [58, 59]. Predictably, the exact urinary bacterial compounds found elevated in ASD do differ in distinct metabolomic studies. This is not surprising since, in addition to differences in sample demographics and sensitivity of available technologies, ethnicity also exerts profound influences on the microbiome, reflecting dietary, genetic, and immunological specificities involved in the host-microbiome interactions [60]. Despite these discrepancies at the level of single compounds, urinary metabolomic studies consistently report an excess of microbiome-derived urinary metabolites, collectively supporting gut dysbiosis in ASD. These results point toward possible negative effects on CNS function exerted by microbiome-derived metabolites. At least three examples are available, albeit with different degrees of support: (a) urinary p-cresol amounts were found correlated with ASD severity [20] or with the intensity of stereotypic behaviors in young autistic children [21]; (b) i.c.v. injection of propionic acid, an enteric-derived short chain fatty acid, produces ASD-like behaviors in the rat [61]; (c) indoxyl sulfate is a known risk factor for cognitive impairment in chronic renal disease [62]: its influx across the blood-brain barrier using the organic anion transporter 3 significantly reduces the efflux of various neurotransmitter metabolites through the same transporter, leading to their accumulation [63]. Importantly, sizable improvements in behavioral and serum metabolome abnormalities were recorded using the maternal immune activation (MIA) rodent model of ASD following the correction of gut dysbiosis using Bacteroides fragilis [64].
Purine metabolites are also well represented in the urines of ASD children, which display a large excess of inosine, hypoxanthine, and xanthosine (Figs. 3 and 5). This pattern bears an interesting resemblance to the excess of urinary inosine and hypoxanthine detected in Fmr1 knock-out mice, an animal model of fragile-X syndrome [35]. Also, mice exposed prenatally to MIA triggered by poly(I:C) injected at E12.5 and E17.5 show an excess of urinary inosine [36]. This excess of urinary purinergic metabolites has been interpreted as part of a “cell danger metabolic response” involving mitochondrial dysfunction, adenosine triphosphate (ATP), and adenosine diphosphate (ADP) release, activation of a variety of purinergic receptors yielding microglial activation, innate, and adaptive immunity responses and leukocyte chemotactics [65]. Inborn errors of purine metabolism are associated with behavioral abnormalities including autistic features [66]. Strikingly, inhibition of purine metabolism by suramin, a competitive antagonist at P2X and P2Y purinergic receptors, reverses behavioral, neurochemical, transcriptional, and metabolomics abnormalities both in the Fmr1 knock-out mouse and in MIA mice exposed to poly(I:C) during pregnancy [35–37]. Conceivably, this metabolic abnormality, shared between human ASD and genetic/immunological rodent models could thus represent a valuable biomarker to help guide therapeutic interventions. In addition, the cell danger response also yields relative vitamin B6 deficiency and the enzyme kynureninase is B6 dependent [65]; hence, a cell danger metabolic response in the presence of adequate tryptophan intake could also explain the decreased kynurenine and increased xanthurenic and quinolinic acid observed here (Fig. 4). Interestingly, these abnormalities have been sometimes overcome with vitamin B6 supplementation [67], a therapeutic approach initially proposed for ASD in conjunction with magnesium supplementation [68]. In light of the present data, B6-Mg++ supplementation in ASD may deserve further scrutiny in urinary biomarker-driven therapeutic trials, as no firm conclusion on its potential efficacy has yet been reached [69].
Conclusions
Targeting young autistic children and tightly matched controls, using the sensitive approach HILIC UHPLC-MS, and applying metabolic pathway analysis, we identified several urinary metabolic pathways significantly altered in ASD: tryptophan, purine, and vitamin B6 metabolisms; phenylalanine, and tyrosine biosynthesis; and to a lesser extent, pantothenate and CoA, riboflavin, and pyrimidine metabolisms. Several of these same pathways, especially tryptophan, purine, and gut microbiome metabolisms, are also abnormal in animal models of ASD and provide very interesting leads toward possible pathophysiological explanations for specific symptoms present in many autistic children, such as seizures and sleep disorders. These metabolic abnormalities may apply to young children only, as suggested by studies of urinary p-cresol [20, 21]. It will indeed be very important to now perform a similar metabolomic assessment on ASD individuals and controls older than 8 years of age. Investigations of CSF metabolomics will be necessary to verify to what extent peripheral results reflect CNS pathophysiology. Finally, studies involving other diagnostic groups bordering with ASD, such as ADHD, intellectual disability, expressive language disorder, and obsessive-compulsive disorder, will be required to assess the disease specificity of the metabolomic abnormalities reported here and to determine their potential value as ASD-specific biomarkers, possibly able to aid clinicians in providing more reliable diagnoses in early infancy.
Abbreviations
- 5-HIAA:
-
5-hydroxyindoleacetic acid
- 5-HT:
-
5-hydroxytryptamine
- 5-HTTP:
-
5-hydroxytryptophan
- AANAT:
-
N-acetylserotonin by arylalkylamine N-acetyltransferase
- ADHD:
-
attention deficit hyperactivity disorder
- ADP:
-
adenosine diphosphate
- ASD:
-
autism spectrum disorder
- ASMT:
-
N-acetylserotonin O-methyltransferase
- ATP:
-
adenosine triphosphate
- CNS:
-
central nervous system
- CSF:
-
cerebrospinal fluid
- ESI:
-
electrospray ionization
- GC:
-
gas chromatography
- HILIC:
-
hydrophilic interaction chromatography
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LC:
-
liquid chromatography
- MetPA:
-
metabolic pathway analysis
- MIA:
-
maternal immune activation
- MS:
-
mass spectroscopy
- NAD:
-
nicotinic acid
- NMDA:
-
N-methyl-d-aspartate
- NMR:
-
nuclear magnetic resonance
- OPLS-DA:
-
orthogonal partial least squares discriminant analysis
- TOF:
-
time of flight
- VIP:
-
variable influence on the projection
References
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Washington, DC; 2013.
Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65:591–8.
Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators, Centers for Disease Control and Prevention (CDC). Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014;63:1–21.
Rutter M. Incidence of autism spectrum disorders: changes over time and their meaning. Acta Paediatr. 2005;94:2–15.
Frans EM, Sandin S, Reichenberg A, Långström N, Lichtenstein P, McGrath JJ, Hultman CM. Autism risk across generations: a population-based study of advancing grandpaternal and paternal age. JAMA Psychiat. 2013;70:516–21.
Persico AM, Merelli S. Environmental factors and autism spectrum disorder. In: Leboyer M, Chaste P, editors. Autism spectrum disorders: phenotypes, mechanisms and treatments. vol. 180. Basilea: Karger; 2015. p. 113–34.
Persico AM, Napolioni V. Autism genetics. Behav Brain Res. 2013;251:95–112.
Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13:247.
Ruggeri B, Sarkans U, Schumann G, Persico AM. Biomarkers in autism spectrum disorder: the old and the new. Psychopharmacology (Berl). 2014;231:1201–16.
Yap IK, Angley M, Veselkov KA, Holmes E, Lindon JC, Nicholson JK. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J Proteome Res. 2010;9:2996–3004.
Ming X, Stein TP, Barnes V, Rhodes N, Guo L. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res. 2012;11:5856–62.
Emond P, Mavel S, Aidoud N, Nadal-Desbarats L, Montigny F, Bonnet-Brilhault F, Barthélémy C, Merten M, Sarda P, Laumonnier F, Vourc’h P, Blasco H, Andres CR. GC-MS-based urine metabolic profiling of autism spectrum disorders. Anal Bioanal Chem. 2013;15:5291–300.
Mavel S, Nadal-Desbarats L, Blasco H, Bonnet-Brilhault F, Barthélémy C, Montigny F, Sarda P, Laumonnier F, Vourc’h P, Andres CR, Emond P. 1H-13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta. 2013;114:95–102.
Nadal-Desbarats L, Aïdoud N, Emond P, Blasco H, Filipiak I, Sarda P, Bonnet-Brilhault F, Mavel S, Andres CR. Combined 1H-NMR and 1H-13C HSQC-NMR to improve urinary screening in autism spectrum disorders. Analyst. 2014;139:3460–8.
Noto A, Fanos V, Barberini L, Grapov D, Fattuoni C, Zaffanello M, Casanova A, Fenu G, De Giacomo A, De Angelis M, Moretti C, Papoff P, Ditonno R, Francavilla R. The urinary metabolomics profile of an Italian autistic children population and their unaffected siblings. J Matern Fetal Neonatal Med. 2014;2:46–52.
Diémé B, Mavel S, Blasco H, Tripi G, Bonnet-Brilhault F, Malvy J, Bocca C, Andres CR, Nadal-Desbarats L, Emond P. Metabolomics study of urine in autism spectrum disorders using a multiplatform analytical methodology. J Proteome Res. 2015;14:5273–82.
West PR, Amaral DG, Bais P, Smith AM, Egnash LA, Ross ME, Palmer JA, Fontaine BR, Conard KR, Corbett BA, Cezar GG, Donley EL, Burrier RE. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS One. 2014;9:e112445.
Nicholson G, Rantalainen M, Li JV, Maher AD, Malmodin D, Ahmadi KR, Faber JH, Barrett A, Min JL, Rayner NW, Toft H, Krestyaninova M, Viksna J, Neogi SG, Dumas ME, Sarkans U, MolPAGE Consortium, Donnelly P, Illig T, Adamski J, Suhre K, Allen M, Zondervan KT, Spector TD, Nicholson JK, Lindon JC, Baunsgaard D, Holmes E, McCarthy MI, Holmes CC. A genome-wide metabolic QTL analysis in Europeans identifies functional effects of two loci shaped by recent positive selection. PLoS Genet. 2011;7:1002270.
Gebregiworgis T, Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb Chem High Throughput Screen. 2012;15:595–610.
Altieri L, Neri C, Sacco R, Curatolo P, Benvenuto A, Muratori F, Santocchi E, Bravaccio C, Lenti C, Saccani M, Rigardetto R, Gandione M, Urbani A, Persico AM. Urinary p-cresol is elevated in small children with autism spectrum disorder. Biomarkers. 2011;16:252–60.
Gabriele S, Sacco R, Cerullo S, Neri C, Urbani A, Tripi G, Malvy J, Barthelemy C, Bonnet-Brihault F, Persico AM. Urinary p-cresol is elevated in young French children with autism spectrum disorder: a replication study. Biomarkers. 2014;19:463–70.
Chugani DC, Muzik O, Behen M, Rothermel R, Janisse JJ, Lee J, Chugani HT. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Ann Neurol. 1999;45:287–95.
Chandana SR, Behen ME, Juhász C, Muzik O, Rothermel RD, Mangner TJ, Chakraborty PK, Chugani HT, Chugani DC. Significance of abnormalities in developmental trajectory and asymmetry of cortical serotonin synthesis in autism. Int J Dev Neurosci. 2005;23:171–82.
Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. Mapping early brain development in autism. Neuron. 2007;56:399–413.
Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am J Psychiatry. 1997;154:185–90.
Hemstrom P, Irgum K. Hydrophilic interaction chromatography. J Sep Sci. 2006;29:1784–821.
Alpert AJ. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 2008;80:62–76.
D’Alessandro A, Gevi F, Zolla L. A robust high resolution reversed-phase HPLC strategy to investigate various metabolic species in different biological models. Mol Biosyst. 2011;7:1024–32.
Gevi F, D’Alessandro A, Rinalducci S, Zolla L. Alterations of red blood cell metabolome during cold liquid storage of erythrocyte concentrates in CPD–SAGM. J Proteomics. 2012;76 Spec No:168–80.
Whiteley P, Waring R, Williams L, Klovrza L, Nolan F, Smith S, Farrow M, Dodou K, Lough WJ, Shattock P. Spot urinary creatinine excretion in pervasive developmental disorders. Pediatr Int. 2006;48:292–7.
Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–26.
Xia J, Mandal R, Sinelnikov I, Broadhurst D, Wishart DS. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–33.
Dettmer K, Aronov PA, Hammock BD. Mass spectrometry based metabolomics. Mass Spectrom Rev. 2007;26:51–78.
Ellis DI, Goodacre R. Metabolic fingerprinting in disease diagnosis: biomedical applications of infrared and Raman spectroscopy. Analyst. 2006;131:875–85.
Naviaux JC, Wang L, Li K, Bright AT, Alaynick WA, Williams KR, Powell SB, Naviaux RK. Antipurinergic therapy corrects the autism-like features in the fragile X (Fmr1 knockout) mouse model. Mol Autism. 2015;6:1.
Naviaux JC, Schuchbauer MA, Li K, Wang L, Risbrough VB, Powell SB, Naviaux RK. Reversal of autism-like behaviors and metabolism in adult mice with single-dose antipurinergic therapy. Transl Psychiatry. 2014;4:e400.
Naviaux RK, Zolkipli Z, Wang L, Nakayama T, Naviaux JC, Le TP, Schuchbauer MA, Rogac M, Tang Q, Dugan LL, Powell SB. Antipurinergic therapy corrects the autism-like features in the poly(IC) mouse model. PLoS One. 2013;8:e57380.
Böhmer C, Bröer A, Munzinger M, Kowalczuk S, Rasko JE, Lang F, Bröer S. Characterization of mouse amino acid transporter B0AT1 (slc6a19). Biochem J. 2005;389(Pt 3):745–51.
Singer D, Camargo SM, Ramadan T, Schäfer M, Mariotta L, Herzog B, Huggel K, Wolfer D, Werner S, Penninger JM, Verrey F. Defective intestinal amino acid absorption in Ace2 null mice. Am J Physiol Gastrointest Liver Physiol. 2012;303:G686–95.
Keszthelyi D, Troost FJ, Masclee AA. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil. 2009;21:1239–49.
Maddison DC, Giorgini F. The kynurenine pathway and neurodegenerative disease. Semin Cell Dev Biol. 2015;40:134–41.
Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2004;57:67–8.
Suzuki K, Sugihara G, Ouchi Y, Nakamura K, Futatsubashi M, Takebayashi K, Yoshihara Y, Omata K, Matsumoto K, Tsuchiya KJ, Iwata Y, Tsujii M, Sugiyama T, Mori N. Microglial activation in young adults with autism spectrum disorder. JAMA Psychiat. 2013;70:49–58.
Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, West AB, Arking DE. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748.
Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, Courchesne E, Everall IP. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68:368–76.
Guillemin G, Smith DG, Smythe GA, Armati PJ, Brew GJ. Expression of the kynurenine pathway enzymes in human microglia and macrophages. Adv Exp Med Biol. 2003;527:105–12.
Guillemin GJ. Quinolinic acid: neurotoxicity. FEBS J. 2012;279:1355.
Kohane IS, McMurry A, Weber G, MacFadden D, Rappaport L, Kunkel L, Bickel J, Wattanasin N, Spence S, Murphy S, Churchill S. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS One. 2012;7:e33224.
Nir I, Meir D, Zilber N, Knobler H, Hadjez J, Lerner Y. Brief report: circadian melatonin, thyroid-stimulating hormone, prolactin, and cortisol levels in serum of young adults with autism. J Autism Dev Disord. 1995;25:641–54.
Kulman G, Lissoni P, Rovelli F, Roselli MG, Brivio F, Sequeri P. Evidence of pineal endocrine hypofunction in autistic children. Neuroendocrinol Lett. 2000;21:31–4.
Tordjman S, Anderson GM, Pichard N, Charbuy H, Touitou Y. Nocturnal excretion of 6-sulphatoxymelatonin in children and adolescents with autistic disorder. Biol Psychiatry. 2005;57:134–8.
Tordjman S, Anderson GM, Bellissant E, Botbol M, Charbuy H, Camus F, Graignic R, Kermarrec S, Fougerou C, Cohen D, Touitou Y. Day and nighttime excretion of 6-sulphatoxymelatonin in adolescents and young adults with autistic disorder. Psychoneuroendocrinology. 2012;37:1990–7.
Melke J, Goubran Botros H, Chaste P, Betancur C, Nygren G, Anckarsäter H, Rastam M, Ståhlberg O, Gillberg IC, Delorme R, Chabane N, Mouren-Simeoni MC, Fauchereau F, Durand CM, Chevalier F, Drouot X, Collet C, Launay JM, Leboyer M, Gillberg C, Bourgeron T. Abnormal melatonin synthesis in autism spectrum disorders. Mol Psychiatry. 2008;13:90–8.
Pagan C, Delorme R, Callebert J, Goubran-Botros H, Amsellem F, Drouot X, Boudebesse C, Le Dudal K, Ngo-Nguyen N, Laouamri H, Gillberg C, Leboyer M, Bourgeron T, Launay JM. The serotonin-N-acetylserotonin–melatonin pathway as a biomarker for autism spectrum disorders. Transl Psychiatry. 2014;4:e479.
Vitte PA, Harthe C, Lestage P, Claustrat B, Bobillier P. Plasma, cerebrospinal fluid, and brain distribution of 14C-melatonin in rat: a biochemical and autoradiographic study. J Pineal Res. 1988;5:437–53.
Rossignol DA, Frye RE. Melatonin in autism spectrum disorders: a systematic review and meta-analysis. Dev Med Child Neurol. 2011;53:783–92.
Bull G, Shattock P, Whiteley P, Anderson R, Groundwater P, Lough JW, Lees G. Indolyl-3- acryloylglycine (IAG) is a putative diagnostic urinary marker for autism spectrum disorders. Med Sci Monit. 2003;9:CR422–5.
Wang L, Angley MT, Gerber JP, Young RL, Abarno DV, McKinnon RA, Sorich MJ. Is urinary indolyl-3-acryloylglycine a biomarker for autism with gastrointestinal symptoms? Biomarkers. 2009;14:596–603.
Wright B, Brzozowski AM, Calvert E, Farnworth H, Goodall DM, Holbrook I, Imrie G, Jordan J, Kelly A, Miles J, Smith R, Town J. Is the presence of urinary indolyl-3-acryloylglycine associated with autism spectrum disorder? Dev Med Child Neurol. 2005;47:190–2.
Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–22.
Thomas RH, Meeking MM, Mepham JR, Tichenoff L, Possmayer F, Liu S, MacFabe DF. The enteric bacterial metabolite propionic acid alters brain and plasma phospholipid molecular species: further development of a rodent model of autism spectrum disorders. J Neuroinflammation. 2012;9:153.
Watanabe K, Watanabe T, Nakayama M. Cerebro-renal interactions: impact of uremic toxins on cognitive function. Neurotoxicology. 2014;44:184–93.
Ohtsuki S, Asaba H, Takanaga H, Deguchi T, Hosoya K, Otagiri M, Terasaki T. Role of blood-brain barrier organic anion transporter 3 (OAT3) in the efflux of indoxyl sulfate, a uremic toxin: its involvement in neurotransmitter metabolite clearance from the brain. J Neurochem. 2002;83:57–66.
Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–63.
Naviaux RK. Metabolic features of the cell danger response. Mitochondrion. 2014;16:7–17.
Nyhan WL, James JA, Teberg AJ, Sweetman L, Nelson LG. A new disorder of purine metabolism with behavioral manifestations. J Pediatr. 1969;74:20–7.
Tada K, Yokoyama Y, Nakagawa H, Yoshida T, Arakawa T. Vitamin B6 dependent xanthurenic aciduria. Tohoku J Exp Med. 1967;93:115–24.
Martineau J, Barthelemy C, Garreau B, Lelord G. Vitamin B6, magnesium, and combined B6-Mg: therapeutic effects in childhood autism. Biol Psychiatry. 1985;20:467–78.
Nye C, Brice A. Combined vitamin B6-magnesium treatment in autism spectrum disorder. Cochrane Database Syst Rev. 2005;4:CD003497.
Acknowledgements
We gratefully acknowledge the patients, controls, and family members who participated in this study and the clinicians who contributed to patient recruitment within the framework of the 2011 collaborative p-cresol project.
Funding
This work was supported by the Italian Ministry for University, Scientific Research and Technology (PRIN n.2006058195), the Italian Ministry of Health (CCM2012), the Fondazione Gaetano e Mafalda Luce (Milan, Italy), the Autism Research Institute (San Diego, CA), and the Innovative Medicines Initiative Joint Undertaking (EU-AIMS, n. 115300). LZ and FG are supported by mobility studentship funds and post-doctoral research grant by the Interuniversity Consortium for Biotechnologies (CIB).
Availability of data and materials
Not applicable.
Authors’ contributions
FG and LZ conducted the metabolomics analyses, analyzed and interpreted the biochemical data, and drafted the manuscript. SG collected the urine samples, interpreted the data, and drafted the manuscript. AMP designed the study, was responsible for clinical assessments, interpreted the data, and wrote the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The Institutional Review Board of University Campus Bio-Medico (Rome, Italy) approved the study protocol (n. 50/15 PAR). All parents gave written informed consent for their children, using the approved consent form.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1: Table S1.
Demographic and clinical characteristics of the autistic sample (N = 30, unless otherwise specified). Typically developing controls were tightly sex- and age-matched, with M:F = 22–8, age 5.03 ± 0.32 years, and no clinical evidence of ASD-related DSM-IV diagnoses or intellectual disability. (DOCX 21 kb)
Additional file 2:
Supplementary methods and references. (DOCX 15 kb)
Additional file 3: Figure S1.
Accurate mass and MS/MS fragmentation data for (a) kynurenine, (b) melatonin, and (c) tryptophan. (TIF 191 kb)
Additional file 5:
Summary statistics. (XLSX 10 kb)
Additional file 6: Figure S2.
Q2 and R2 data pertaining to the PCA. *p < 0.05 refers to the best values of the currently selected measures (Q2). (TIF 233 kb)
Additional file 7: Figure S3.
ROC curve for the top 25 most discriminating metabolites between ASD cases and controls, displayed in Fig. 2. (TIF 131 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gevi, F., Zolla, L., Gabriele, S. et al. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Molecular Autism 7, 47 (2016). https://doi.org/10.1186/s13229-016-0109-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13229-016-0109-5