Abstract
Background
Analyzing close species with diverse developmental modes is instrumental for investigating the evolutionary significance of physiological, anatomical and behavioral features at a molecular level. Many examples of trait loss are known in metazoan populations living in dark environments. Tunicates are the closest living relatives of vertebrates and typically present a lifecycle with distinct motile larval and sessile adult stages. The nervous system of the motile larva contains melanized cells associated with geotactic and light-sensing organs. It has been suggested that these are homologous to vertebrate neural crest-derived melanocytes. Probably due to ecological adaptation to distinct habitats, several species of tunicates in the Molgulidae family have tailless (anural) larvae that fail to develop sensory organ-associated melanocytes. Here we studied the evolution of Tyrosinase family genes, indispensible for melanogenesis, in the anural, unpigmented Molgula occulta and in the tailed, pigmented Molgula oculata by using phylogenetic, developmental and molecular approaches.
Results
We performed an evolutionary reconstruction of the tunicate Tyrosinase gene family: in particular, we found that M. oculata possesses genes predicted to encode one Tyrosinase (Tyr) and three Tyrosinase-related proteins (Tyrps) while M. occulta has only Tyr and Tyrp.a pseudogenes that are not likely to encode functional proteins. Analysis of Tyr sequences from various M. occulta individuals indicates that different alleles independently acquired frameshifting short indels and/or larger mobile genetic element insertions, resulting in pseudogenization of the Tyr locus. In M. oculata, Tyr is expressed in presumptive pigment cell precursors as in the model tunicate Ciona robusta. Furthermore, a M. oculata Tyr reporter gene construct was active in the pigment cell precursors of C. robusta embryos, hinting at conservation of the regulatory network underlying Tyr expression in tunicates. In contrast, we did not observe any expression of the Tyr pseudogene in M. occulta embryos. Similarly, M. occulta Tyr allele expression was not rescued in pigmented interspecific M. occulta × M. oculata hybrid embryos, suggesting deleterious mutations also to its cis-regulatory sequences. However, in situ hybridization for transcripts from the M. occulta Tyrp.a pseudogene revealed its expression in vestigial pigment cell precursors in this species.
Conclusions
We reveal a complex evolutionary history of the melanogenesis pathway in tunicates, characterized by distinct gene duplication and loss events. Our expression and molecular data support a tight correlation between pseudogenization of Tyrosinase family members and the absence of pigmentation in the immotile larvae of M. occulta. These results suggest that relaxation of purifying selection has resulted in the loss of sensory organ-associated melanocytes and core genes in the melanogenesis biosynthetic pathway in M. occulta.
Similar content being viewed by others
Background
Closely related species with different modes of development are attractive systems to investigate the molecular bases of evolutionary changes. The evolutionary loss of physiological, anatomical and behavioral traits by cave-dwelling animals is one noteworthy example [1]. For instance, several studies have revealed convergent or parallel evolution of loss of pigmentation, eyes and various behavioral adaptations in various populations of the Mexican cave tetra (Astyanax mexicanus), which have evolved in parallel from surface-dwelling populations [2,3,4]. Insights into similar processes in cave-dwelling crustaceans are beginning to emerge [5].
Tunicates are the sister group to the vertebrates and are characterized by a life cycle with distinct motile larval and sessile adult stages [6, 7]. The tailed, tadpole-type larva of a typical tunicate consists of a head containing a brain, and a tail containing a dorsal nerve cord and a notochord flanked by striated muscle cells. Larvae swim by alternated left–right contractions of the tail muscles, which are controlled by a minimal central nervous system containing only 177 neurons [8]. Inside the brain, two sensory organs containing a single melanocyte each are generally present: the otolith, for gravity perception, and the ocellus, for the detection of light [9, 10]. These two distinct pigment cells develop from a single bilateral pair of cells along the neural plate borders [11] and are proposed to be homologous to pigment cells of the dorsal ocelli in amphioxus [12, 13] and to neural crest-derived melanocytes in vertebrates [14, 15]. Pigmentation in tunicates depends critically on the activity of melanogenic enzymes Tyrosinase (Tyr) and Tyrosinase-related proteins (Tyrps) [16, 17], as is the case in vertebrates [18].
Despite the deeply conserved body plan of the swimming tunicate larva, some species instead develop a tailless (anural) larva which is immotile [19,20,21]. About 20 tunicate species have been shown to exhibit anural development, and almost all of these species are members of the family Molgulidae, in which tail loss has occurred independently multiple times [21, 22]. The ecological significance of these losses is still not clear, although it has been suggested that this could be related to the habitats to which these species are adapted, notably sand flats and rocky shores at higher latitudes [22].
Anural tunicate larvae have conspicuously lost most of those features more directly associated with swimming. For instance, they have lost the ability to generate differentiated tail muscles [23,24,25,26,27,28], and produce rounded larvae without the characteristic elongated, striated tail muscles typical of most tunicate larvae. The anural species Molgula occulta has lost genes important for muscle function such as Muscle actin, which has become pseudogenized by deletions, insertions and codon substitutions in coding and noncoding regions, resulting in truncated proteins and/or reduced transcriptional activity [29, 30]. However, tail muscles are still specified in anural embryos, which express the same regulatory cascade for myogenesis in tailed species [31, 32]. Similarly, anural embryos also specify notochord cells, but these later fail to undergo proper morphogenesis by convergent extension [33].
In contrast the loss of functional muscles and notochord, the loss of differentiated neural structures in anural larvae has not been studied in detail. The most obvious evolutionary loss of a neural structure is that of the sensory pigment cells, or melanocytes. While tunicate larvae generally possess both an ocellus and otolith, most molgulids have only an otolith. They have lost the ocellus and, concomitantly, the ability to respond to light [34]. In Molgula species with anural larvae, there has been a more complete loss of pigmentation, as even the otolith is missing [19] or vestigial [26]. This has been postulated to be due the lack of adaptive value of the otolith for such anural larvae, which do not swim and therefore do not need to sense their orientation in the water column as swimming, tailed larvae do [22].
Here we describe the development of sensory pigment cells in the embryos of Molgula oculata, and the molecular basis of pigmentation loss in the closely related, anural and unpigmented species M. occulta. We found that inactivated Tyrosinase (Tyr) and Tyrosinase-related protein (Tyrp) genes are present in the genome of M. occulta, but are not likely to produce functional enzymes. While Tyr transcripts were not detected in M. occulta embryos, transcription of a Tyrp gene was observed in vestigial pigment cells. Surprisingly, more than two distinct Tyrosinase alleles with different inactivating mutations were cloned from a sampling of M. occulta individuals, suggesting parallel loss of pigmentation by independent loss-of-function mutations Tyrosinase within the same population.
Results
Tyrosinase family enzymes in Molgula
The melanogenic toolkit of tunicates and vertebrates revolves around Tyrosinase family gene members, which encode the rate-limiting enzymes that catalyze the final steps in melanin biosynthesis [16]. In Ciona spp., the family is comprised by Tyrosinase (Tyr), Tyrosinase-related protein a (Tyrp.a) and Tyrosinase-related protein b (Tyrp.b). We searched for orthologs of these genes by performing tBlastn [35] on the recently published genomes of tailless, unpigmented species Molgula occulta and the closely related, tailed, pigmented species Molgula oculata [36], using the Ciona robusta (formerly known as Ciona intestinalis type A) [37] and Halocynthia roretzi [38] orthologs as query sequences.
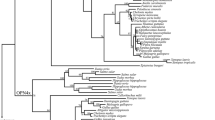
We found partial sequences for one Tyr (Mooccu.Tyr) and one Tyrp (Mooccu.Tyrp.a) ortholog in the genome of M. occulta. In contrast, we were able to identify a single Tyr ortholog (Moocul.Tyr) and at least four distinct Tyrp paralogs in the genome of the M. oculata (Moocul.Tyrp.a, Moocul.Tyrp.b, Moocul.Tyrp.c and Moocul.Tyrp.d). Previously, phylogenetic analyses had shown that Ciona Tyrp.a and Tyrp.b are the result of a tunicate-specific independent duplication of an ancestral Tyrp gene, equally related to the vertebrate paralogs Tyrp1 and Tyrp2 (Dopachrome tautomerase) [16]. An ascidian phylogenetic tree encompassing C. robusta, Ciona savignyi and Phallusia mammillata for Phlebobranchia, Halocynthia roretzi, Botryllus schlosseri, M. occulta, M. oculata and M. occidentalis for Stolidobranchia sequences does not support one-to-one orthology between any of the Ciona and Molgula Tyrp genes (Fig. 1), suggesting that a single ancestral Tyrp gene likely underwent independent duplications in different tunicate clades.
Tyrosinase family evolution in tunicates. Maximum likelihood phylogeny of Tyrosinase members available in databases: The numbers at the branches represent the replicates come out employing ML method. Two distinct colored boxes have been used to highlight Tyr (blue box) and Tyrp (pink box) protein classes
We also searched for Tyr and Tyrp mRNA transcripts in the transcriptomes obtained from embryos of both species at three different developmental stages (late gastrula, neurula and tailbud) [39]. Sequences from all Tyr family genes from M. oculata were identified in the corresponding transcriptome, suggesting that all these genes are expressed during embryogenesis of pigmented Molgula species. By contrast, only Mooccu.Tyrp.a was present in the M. occulta embryonic transcriptome, hinting at the possibility that the melanogenic toolkit is only partially deployed during the development of this species’ unpigmented, anural larvae. Furthermore, Mooccu.Tyr transcripts were undetectable by RNAseq profiling of interspecies hybrid embryos, resulting from fertilization of M. oculata eggs with M. occulta sperm [39]. While 70–215 RNAseq reads could be mapped to the M. oculata Tyr allele in hybrid embryos at each developmental stage, only one or two reads mapped to M. occulta Tyr alleles at any stage (see Table in Additional file 1). Thus, even in embryos actively transcribing the M. oculata Tyr allele at high levels, the M. occulta allele is largely transcriptionally inactive. Taken together, these data indicate that the M. occulta Tyr gene is transcriptionally silent during embryonic development. Since hybrid embryos have pigmented cells [33], our data also indicate that this rescue of melanization in the interspecific hybrids is due entirely to the M. oculata Tyrosinase allele.
Pigment cell marker expression in Molgula embryos
We next sought to determine the spatiotemporal expression pattern of selected M. occulta and M. oculata Tyrosinase family genes, by whole-mount mRNA in situ hybridization on embryos at different developmental stages (Fig. 2). In the larvae of M. occulta, bilateral Mooccu.Tyrp.a expression was detected in embryos starting at 10 h post-fertilization (hpf), which is around the time they are hatching (Fig. 2a). Their number and location in the embryo suggest they correspond to vestigial pigment cell precursors (a9.49 lineage in Ciona robusta, Fig. 2b). In contrast, Mooccu.Tyr is absent from the transcriptome (see above) [39]. Indeed, mRNA in situ hybridization using a riboprobe prepared from a genomic clone of the Mooccu.Tyr locus (see below) showed no transcriptional activity of this sequence during development nor in hatched larvae (Fig. 2d, e). This further confirms the complete loss of transcriptional activity of this gene in M. occulta as suggested by the RNAseq data. Taken together, these data suggest that, although unpigmented M. occulta specify vestigial pigment cell precursors that transcribe Mooccu.Tyrp.a, the rate-limiting melanogenic gene Mooccu.Tyr is not expressed in these cells.
Expression of Tyrosinase genes during tunicate embryogenesis. The whole-mount in situ hybridization experiments show the localization of Tyrosinase gene expression in pigment cell precursors of three tunicates: Mooccu.Tyrp.a in Molgula occulta hatched larva (a), Cirobu.Tyr in Ciona robusta neurula (b), Moocul.Tyr in Molgula oculata neurula embryos (c). Double in situ hybridization of M. occulta Tyrosinase and either Neurogenin (d) or Onecut (e), showing lack of Tyrosinase expression in the developing central nervous system of embryos and larvae at various stages
To verify whether the Tyrosinase ortholog is expressed in the a9.49 lineage cells of the pigmented species M. oculata, we performed in situ hybridization on embryos of this species. At 7 hpf, corresponding to neurula stage, Moocul.Tyr is expressed in two cells in the neural plate borders corresponding to the pigment cell precursors (Fig. 2c). Later in development at 10 hpf, Moocul.Tyr is expressed around the melanizing otolith (see Figure in Additional file 1). We also isolated an intergenic sequence of ~1.3 kbp upstream of the coding ATG in the Moocul.Tyr gene and cloned this region in a reporter construct containing GFP (Moocul.Tyr > GFP). This reporter was tested by electroporation into the pigmented embryos of the distantly related Molgula occidentalis, the only Molgula species that can be easily transfected with plasmid DNA, and in C. robusta. In both species, GFP expression was detected in four cells corresponding to the putative pigment cell lineage in the central nervous system of the tailbud embryo (Fig. 3).
GFP reporter assay of Molgula oculata Tyr promoter. A plasmid containing GFP has been used to demonstrate the conservation of Moocul.Tyr regulatory activity (1.3 Kb fragment) by electroporation of tunicate embryos. GFP expression is detected in the pigment cell lineage of tailbud-stage embryos of Molgula occidentalis (left) and Ciona robusta (right)
The four cells seen in transgenic Ciona and M. occidentalis embryos expressing M. oculata Tyr > GFP reporter are the presumptive ocellus and otolith pigment cells along with their more anterior sister cells that express Tyr transiently [40], all converged to form a single row at the midline of the embryo. As mentioned previously, tailed Molgula species like M. oculata and M. occidentalis lack an ocellus and only produce an otolith pigment cell. However, from these data we conclude that in Molgula and Ciona, both presumptive ocellus and otolith pigment cells are specified early and express Tyr/Tyrp.a. Thus, the gene regulatory network for pigment cell specification and Tyr activation appears to be conserved between Molgula and Ciona, but ocellus differentiation in Molgula is somehow suppressed after pigment cell specification.
Tyrosinase family pseudogenes in M. occulta
The lack of melanin production in M. occulta pigment cell lineage hinted at the possibility that one or more Tyrosinase family genes have become inactivated in this species, through mutations to coding and/or noncoding sequences. While Mooccu.Tyr does not appear to be expressed, transcripts for Mooccu.Tyrp.a are still detected. However, this does not necessarily indicate the presence of functional Tyrp.a protein. A precedent for this has been found in the Muscle actin genes of M. occulta, which have been inactivated by mutations that affect both their cis-regulatory and protein-coding sequences [30].
The alignment of the M. occulta and M. oculata Tyrp.a sequences from the assembled genomes [36] revealed the presence of a 4-bp insertion in Mooccu.Tyrp.a, resulting in a frameshift and a premature stop codon in the middle of the gene, right at the start of the conserved Tyrosinase domain which contains the active site of the enzyme (Fig. 4a). This indicates that Mooccu.Tyrp.a transcripts do not code for a full-length, functional enzyme.
Tyrosinase family pseudogenization in Molgula occulta. The figure points out the presence of premature stop codons (red) and frameshift insertions (magenta) in genomic sequences of Tyrp.a (a) and Tyr (b). A reconstruction of the active site of the proteins has been obtained using the SMART Web site [58]. Certain Mooccu.Tyr alleles are characterized by the presence of transposable elements (yellow) inserted just upstream of the active site (b)
We also identified a scaffold from the available genome assembly that includes part of the Mooccu.Tyr gene, with an assembly gap in the middle of the protein-coding region. To circumvent this assembly problem and analyze the Mooccu.Tyr gene in greater detail, we sought to clone this sequence from genomic DNA extracted from several individuals. To this end, we extracted the genomic DNA from ten individuals and amplified the region from each sample by PCR. This resulted unexpectedly in some individuals carrying Tyr alleles of different lengths, suggesting the presence of one or more structural variants of the same Tyr gene in the population. Each unique PCR product was purified and cloned into plasmids. When the resulting clones were sequenced, we identified three distinct Mooccu.Tyr alleles (Fig. 4b). In the portion of the coding sequence analyzed, one of the alleles (NP2) has a 5-bp frameshifting insertion that results in a premature stop codon, in the beginning of the conserved Tyrosinase domain and enzyme active site (Fig. 4b). The two other alleles (NP1 and NY1) each have a large, ~460-bp insertions with terminal inverted repeats, bearing the hallmarks of a transposable element. These large insertions have several in-frame stop codons. Curiously, NP1 and NY1 have the same element, but in opposite orientations, suggesting one configuration might be derived from the other by a re-transposition event (see Additional file 2). NP1 and NY1 also have a shorter frameshifting insertion downstream of the transposable element, also in the beginning of the Tyrosinase domain, distinct from that of NP2, suggesting a possible independent origin from the functional common ancestor (Fig. 4b).
Discussion
Melanocytes and related pigment cells share some basic characteristics: Melanin pigments are synthesized by Tyrosinase and Tyrosinase-related enzymes [41] and stored in melanosomes, which are tissue-specific lysosome-related organelles distributed in the cell by a regulated vesicular trafficking system. Loss of pigmentation in humans and other vertebrates can be caused by mutations in Tyrosinase itself, or other genes encoding proteins important for substrate and/or pigment transport and storage [42, 43]. Our phylogenetic survey pointed at a complex evolutionary history for Tyrosinase family in ascidians, with a pattern influenced by duplications and losses (Fig. 1). In particular, we discovered a different number of Tyrp paralogs in Molgula spp., suggesting the occurrence of many duplicative events in distinct tunicate clades followed, in some cases, by gene losses. It would be interesting to comprehend the evolutionary and functional relevance of these genomic events. It has been recently showed that tunicate melanocyte specification depends on the FGF/MAPK/Ets pathway, which renders the lineage competent to respond to a Wnt signal [44]. Characterization of the transcriptional response to FGF signaling in the pigment cell lineage has allowed us to identify its molecular mechanisms underlying pigment cell fate choice and, more broadly, anterior–posterior patterning of Ciona central nervous system [40]. It will be interesting to assess how these events are regulated in Molgula species, given that only one pigment cell (the otolith) differentiates.
It was previously suggested that Tyrosinase activity is present in M. occulta juveniles, in spite of a lack of discernible pigmentation in juveniles or adults [45]. Here we show that genes coding for key enzymatic components of the melanogenesis pathway, namely Tyrosinase and Tyrp.a, have been mutated in M. occulta and are highly unlikely to code for functional enzymes, calling into question how Tyrosinase enzyme activity could be maintained in this species. Furthermore, assembled genome sequences of three Molgula species have not uncovered any cryptic Tyr paralogs that could compensate for these loss-of-function mutations. In Ciona, a single Tyr gene is necessary for melanogenesis and sensory organs are unpigmented in larvae carrying Tyr loss-of-function mutations [17]. Our results suggest that the loss of this melanogenic toolkit underlies lack of pigmentation in M. occulta and contradicts the alternate hypothesis that the toolkit is present, but the pigment cells are not specified [45].
The identification of three distinct, inactivated alleles of Mooccu.Tyr in such a small sample of individuals from the same population suggests pigmentation loss in this species has occurred through accumulation of deleterious mutations, which would indicate a relaxation of the purifying selection that maintains a functional melanogenesis toolkit in species with swimming larvae. This agrees with the suggestion that the ability to detect larval orientation by means of the pigmented otolith is of little adaptive value to the immotile larvae of M. occulta [22]. Since melanin appears to be present only in the otolith, and no other larval or adult tissue, the melanogenesis toolkit would have been dispensable for species with anural larvae such as M. occulta.
Furthermore, Mooccu.Tyrp.a is predicted to code for a truncated protein, but is still transcribed in the pigment cell lineage, indicating that it still retains functional cis-regulatory sequences. This is in contrast to Mooccu.Tyr, which is not expressed (even in interspecific hybrids) and thus is likely to bear inactivating mutations in its cis-regulatory sequences as well (Fig. 5). This diversity of mutations suggests that loss of melanogenesis pathway genes is still an ongoing process in M. occulta populations. Loss of pigmentation in different populations of Astyanax cavefish can occur through independent mutations in the same gene [4]. In contrast, loss of pigmentation in the cave crustacean Asellus aquaticus can occur through mutations in different genes in the same population [5]. Our discovery of pigment loss in M. occulta through independent mutations in the same gene in the same population is thus a distinct variation on this theme. It will be interesting to understand how population structure and the genetic architecture of pigmentation in Molgula may have contributed to this.
Conclusions
This study shed light on the molecular mechanisms underlying loss of pigmentation in the annual larvae of Molgula occulta (Fig. 5). Our data point to a role of independent mutations in at least two Tyrosinase family genes in the same population. These mutations are highly likely to result in enzymatically inactive Tyr and Tyrp proteins, which would explain pigment loss in M. occulta, even though we show this species still specifies pigment cell precursors like its pigmented relatives. The identification of several seemingly independent loss-of-function Tyr mutations in the M. occulta population suggests a relaxation of purifying selection may have contributed to loss of pigment cells in this species.
Methods
Phylogenetic analysis
In phylogenetic reconstruction, we employed sequences that have been retrieved from the NCBI (http://www.ncbi.nlm.nih.gov), Ensembl (http://www.ensembl.org/index.html) and Aniseed (http://www.aniseed.cnrs.fr/) [46] databases excluding uninformative proteins (see Additional file 3). Ciona robusta Tyr and Tyrp.a were the initial query sequences used for TBLASTN [35] in other tunicates, and reciprocal blasts were performed on every genome/transcriptome [36, 46, 47]. The protein set was aligned by ClustalW with default parameters [48]. The phylogenetic tree inference was computed employing the maximum likelihood estimation (MLE) using MEGA6 with the WAG + γ + I matrix [49], in order to improve sensitiveness in our analysis. We assessed the robustness of tree topologies with 1000 bootstrap replicates, and the numbers at the branches indicate replicates obtained using the maximum likelihood estimation method (Fig. 1). The graphical representation of trees was constructed using Dendroscope [50].
M. occulta × M. oculata hybrid embryo transcriptome analysis
Reads from RNAseq of M. occulta × M. oculata hybrid embryos from three different stages, gastrula, neurula and tailbud [39], were mapped to the three M. occulta Tyr allele sequences cloned, or the M. oculata Tyr transcript sequence, using Salmon v0.6.0 [51].
Molecular cloning
Putative orthologs of Ciona robusta and Halocynthia roretzi protein-coding genes were initially identified by TBLASTN. Identified sequences were aligned to each other to support orthology relationships within Molgula and then queried by BLASTP against the NCBI nonredundant protein sequence database to further support orthology relationships to well-known tunicate and vertebrate proteins. The cDNAs were cloned by RT-PCR, and the template cDNA libraries were prepared from total RNA extracted from embryos of various stages. Upstream regulatory sequences were cloned by PCR from genomic DNA (see Additional file 4). For PCRs, we used Phusion high-fidelity polymerase (New England Biolabs, Ipswich, MA) or Pfx Platinum polymerase (Thermo Fisher). Genes are named according to the proposed unified nomenclature system for tunicate genetic elements [52]. For genotyping NY1 Tyr allele, primers Moocc.genTyr1 FW and Moocc.genTyr1 REV were used. For genotyping NP1 and NP2 Tyr alleles, primers Moocc.genTyr2 FW and Moocc.genTyr2 REV were used. All the employed oligos are listed in Additional file 5.
In situ hybridization
In situ hybridization assays were performed as previously described, with modifications [53,54,55]. Embryos were fixed in MEM-3.2 to 4% paraformaldehyde buffers for at least 2 h. Pre-hybridization proteinase K concentrations ranged from 0.25 (110 cell stage) to 1 (tailbud) and 5 μg/ml (larvae) for Molgula spp. embryos. The Ciona robusta Tyr and Tyrp.a probe was synthesized from template plasmid from the Gene Collection Release 1 [56].
Probe for fluorescent mRNA in situ hybridization of M. occulta Tyr was prepared from genomic clone NY1, since this gene is not transcriptionally active, and thus, no cDNA could be cloned. Probes for fluorescent mRNA in situ hybridization of M. occulta Neurogenin and Onecut were prepared from cDNA (Additional file 4). Double- and single-color fluorescent in situ hybridization was performed as previously described [53, 55].
Sample collection and electroporation of plasmid DNA into embryos
Gravid adults of M. oculata and M. occulta were collected at the Station Biologique in Roscoff, France, during August (the only time of year when gravid animals can be found). Gonads were dissected to obtain gametes from both species. Gametes were fertilized with conspecific sperm, and embryos were allowed to develop to the appropriate stage and were then dechorionated as described previously [36]. M. occidentalis gravid adults were obtained from Gulf Specimen Marine Lab. Animal handling and electroporation were carried out as described in Stolfi et al. [36]. C. robusta (intestinalis ‘Type A’) adults were collected in San Diego, CA (M-Rep), and in the gulf of Taranto, Italy. Electroporation has been carried out as described [57].
References
Retaux S, Casane D. Evolution of eye development in the darkness of caves: adaptation, drift, or both? Evodevo. 2013;4(1):26.
Kowalko JE, Rohner N, Rompani SB, Peterson BK, Linden TA, Yoshizawa M, Kay EH, Weber J, Hoekstra HE, Jeffery WR, et al. Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr Biol. 2013;23(19):1874–83.
McGaugh SE, Gross JB, Aken B, Blin M, Borowsky R, Chalopin D, Hinaux H, Jeffery WR, Keene A, Ma L, et al. The cavefish genome reveals candidate genes for eye loss. Nat Commun. 2014;5:5307.
Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38(1):107–11.
Protas ME, Trontelj P, Patel NH. Genetic basis of eye and pigment loss in the cave crustacean, Asellus aquaticus. Proc Natl Acad Sci USA. 2011;108(14):5702–7.
Jeffery AK, Swalla BJ. Tunicates. In: Gilbert SF, Raunio AM, editors. Embryology, constructing the organism. London: Elsevier; 1997.
Satoh N. Developmental biology of ascidians. Cambridge: Cambridge University Press; 1994.
Ryan K, Lu Z, Meinertzhagen IA. The CNS connectome of a tadpole larva of Ciona intestinalis (L) highlights sidedness in the brain of a chordate sibling. Elife. 2016;5:1.
Dilly PN. Studies on the receptors in the cerebral vesicle of the ascidian tadpole. I. The otolith. Q J Microsc Sci. 1962;103:393–8.
Tsuda M, Sakurai D, Goda M. Direct evidence for the role of pigment cells in the brain of ascidian larvae by laser ablation. J Exp Biol. 2003;206(8):1409–17.
Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol. 1987;121(2):526–41.
Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature. 2015;527(7578):371–4.
Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 2008;18(7):1127–32.
Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492(7427):104–7.
Wada H, Holland PW, Sato S, Yamamoto H, Satoh N. Neural tube is partially dorsalized by overexpression of HrPax-37: the ascidian homologue of Pax-3 and Pax-7. Dev Biol. 1997;187(2):240–52.
Esposito R, D’Aniello S, Squarzoni P, Pezzotti MR, Ristoratore F, Spagnuolo A. New insights into the evolution of metazoan tyrosinase gene family. PLOS ONE. 2012;7(4):e35731.
Jiang D, Tresser JW, Horie T, Tsuda M, Smith WC. Pigmentation in the sensory organs of the ascidian larva is essential for normal behavior. J Exp Biol. 2005;208(3):433–8.
Tief K, Hahne M, Schmidt A, Beermann F. Tyrosinase, the key enzyme in melanin synthesis, is expressed in murine brain. Eur J Biochem. 1996;241(1):12–6.
Berrill NJ. Studies in tunicate development. Part II. Abbreviation of development in the Molgulidae. Philos Trans R Soc B Biol Sci. 1931;219:281–346.
Lacaze-Duthiers H. Histoire des ascidies simples des cotes de France: 2: Etudes des especes; 1874.
Jeffery WR, Swalla BJ. The myoplasm of ascidian eggs: a localized cytoskeletal domain with multiple roles in embryonic development. Semin Cell Biol. 1990;1(5):373–81.
Huber JL, da Silva KB, Bates WR, Swalla BJ. The evolution of anural larvae in molgulid ascidians. Semin Cell Dev Biol. 2000;11(6):419–26.
Bates WR. Direct development in the Ascidian Molgula-retortiformis (Verrill, 1871). Biol Bull. 1995;188(1):16–22.
Bates WR, Mallett JE. Ultrastructural and histochemical-study of anural development in the ascidian Molgula-pacifica (Huntsman). Roux Arch Dev Biol. 1991;200(4):193–201.
Swalla BJ, Jeffery WR. Modification of the myoplasm in eggs of anural ascidians. Am Zool. 1990;30(4):A42.
Swalla BJ, Jeffery WR. Vestigial brain melanocyte development during embryogenesis of an anural ascidian. Dev Growth Differ. 1992;34(1):17–25.
Tagawa K, Jeffery WR, Satoh N. The recently-described ascidian species Molgula tectiformis is a direct developer. Zool Sci. 1997;14(2):297–303.
Whittaker JR. Development of vestigial tail muscle acetylcholinesterase in embryos of an anural ascidian species. Biol Bull. 1979;156(3):393–407.
Jeffery WR, Swalla BJ, Ewing N, Kusakabe T. Evolution of the ascidian anural larva: evidence from embryos and molecules. Mol Biol Evol. 1999;16(5):646–54.
Kusakabe T, Swalla BJ, Satoh N, Jeffery WR. Mechanism of an evolutionary change in muscle cell differentiation in ascidians with different modes of development. Dev Biol. 1996;174(2):379–92.
Gyoja F, Satou Y, Shin-i T, Kohara Y, Swalla BJ, Satoh N. Analysis of large scale expression sequenced tags (ESTs) from the anural ascidian, Molgula tectiformis. Dev Biol. 2007;307(2):460–82.
Takada N, Satoh N, Swalla BJ. Expression of Tbx6, a muscle lineage T-box gene, in the tailless embryo of the ascidian Molgula tectiformis. Dev Genes Evol. 2002;212(7):354–6.
Swalla BJ, Jeffery WR. Interspecific hybridization between an anural and urodele ascidian: differential expression of urodele features suggests multiple mechanisms control anural development. Dev Biol. 1990;142(2):319–34.
Grave C. Molgula citrina (Alder and Hancock). Activities and structure of the free-swimming larva. J Morphol. 1926;42:453–71.
Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41.
Stolfi A, Lowe EK, Racioppi C, Ristoratore F, Brown CT, Swalla BJ, Christiaen L. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. eLife. 2014. doi:10.7554/eLife.03728.
Brunetti R, Gissi C, Pennati R, Caicci F, Gasparini F, Manni L. Morphological evidence that the molecularly determined Ciona intestinalis type A and type B are different species: Ciona robusta and Ciona intestinalis. J Zool Syst Evol Res. 2015;53(3):186–93.
Sato S, Masuya H, Numakunai T, Satoh N, Ikeo K, Gojobori T, Tamura K, Ide H, Takeuchi T, Yamamoto H. Ascidian tyrosinase gene: its unique structure and expression in the developing brain. Dev Dyn. 1997;208(3):363–74.
Lowe E, Swalla BJ, Brown CT. Evaluating a lightweight transcriptome assembly pipeline on two closely related ascidian species. PeerJ Prepr. 2014;2:e505v1.
Racioppi C, Kamal AK, Razy-Krajka F, Gambardella G, Zanetti L, di Bernardo D, Sanges R, Christiaen LA, Ristoratore F. Fibroblast growth factor signalling controls nervous system patterning and pigment cell formation in Ciona intestinalis. Nat Commun. 2014;5:4830.
Kobayashi T, Urabe K, Winder A, Jimenez-Cervantes C, Imokawa G, Brewington T, Solano F, Garcia-Borron JC, Hearing VJ. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 1994;13(24):5818–25.
Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, Jiang Y, Bittner M, Hammer JA 3rd, Pavan WJ. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci USA. 2002;99(7):4471–6.
Oiso N, Riddle SR, Serikawa T, Kuramoto T, Spritz RA. The rat Ruby (R) locus is Rab38: identical mutations in Fawn-hooded and Tester-Moriyama rats derived from an ancestral Long Evans rat sub-strain. Mamm Genome. 2004;15(4):307–14.
Squarzoni P, Parveen F, Zanetti L, Ristoratore F, Spagnuolo A. FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development. 2011;138(7):1421–32.
Jeffery WR. Evolution and development of brain sensory organs in molgulid ascidians. Evol Dev. 2004;6(3):170–9.
Brozovic M, Martin C, Dantec C, Dauga D, Mendez M, Simion P, Percher M, Laporte B, Scornavacca C, Di Gregorio A. ANISEED 2015: a digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. 2015; 44:D808–18.
Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298(5601):2157.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61(6):1061–7.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;4:417–9.
Stolfi A, Sasakura Y, Chalopin D, Satou Y, Christiaen L, Dantec C, Endo T, Naville M, Nishida H, Swalla BJ, et al. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis. 2015;53(1):1–14.
Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134(18):3297–305.
Christiaen L, Wagner E, Shi W, Levine M. Whole-mount in situ hybridization on sea squirt (Ciona intestinalis) embryos. Cold Spring Harb Protoc. 2009;2009(12):pdb prot5348.
Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain-hindbrain boundary region. Dev Biol. 2007;312(2):631–43.
Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, et al. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33(4):153–4.
Christiaen L, Wagner E, Shi W, Levine L. Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harb Protoc. 2009;12:pdb.prot5345.
Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2014;43(D1):D257–60.
Authors’ contributions
CR collected and fixed samples, CR, MV, UC performed in situ hybridization analysis, AS, MV, UC performed genomic and transgenic analysis, EL and TB performed bioinformatics analysis, LC, BS, MR conceived the experiments, collected and analyzed data, and CR, AS, UC and FR wrote the manuscript. All the authors critically read the manuscript. All authors read and approved the final manuscript
Acknowledgements
We thank the Station Biologique faculty and staff, especially Stéphane Hourdez, Sophie Booker, and Xavier Bailly for their help in carrying out our studies at Roscoff. We thank Daniel J. Richter for comments and advice on the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its additional files).
Funding
The work has been supported by ASSEMBLE MARINE for MoEvoDevo project. CR has been supported by a long-term fellowship ALTF 1608-2014 from EMBO and by a travel grant from the Boehringer Ingelheim Fonds and ASSEMBLE (Association of European Marine Biological Laboratories). AS is supported by NIH award K99 HD084814. UC is supported by a Ph.D. Open University fellowship from SZN. This material is also based in part upon collaborative work by BJS, CTB and EKL supported by the National Science Foundation under Cooperative Agreement No. DBI-0939454 BEACON, A Center for the Study of Evolution in Action. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
13227_2017_74_MOESM1_ESM.pdf
Additional file 1. Figure A: Whole-mount in situ hybridization of Moocul.Tyr in Molgula oculata showing gene expression in one pigment cell. Table: RNAseq read counts from interspecific (M. occulta × M. oculata) embryos mapped to putative parental alleles, showing expression of M. oculata allele but not M. occulta.
13227_2017_74_MOESM3_ESM.txt
Additional file 3. Database of protein sequences used in phylogenetic survey in Fig. 1, obtained from public databases (NCBI, Ensembl, Aniseed).
13227_2017_74_MOESM4_ESM.docx
Additional file 4. Database of cloned sequences that have been used for in situ hybridizations and electroporation experiments.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Racioppi, C., Valoroso, M.C., Coppola, U. et al. Evolutionary loss of melanogenesis in the tunicate Molgula occulta . EvoDevo 8, 11 (2017). https://doi.org/10.1186/s13227-017-0074-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13227-017-0074-x