Abstract
Purpose
Based on the recycling principle, returning of tobacco crop residues into the field is a common agronomic practice. However, comprehensive knowledge about the effects of tobacco plant residue return on the rhizosphere soil microbial community is very limited.
Methods
After tobacco crop residue returning into the potted soil, 16S ribosomal RNA (rRNA) and internal transcribed spacer (ITS) amplicon sequencing were employed to investigate the bacterial and fungal communities, respectively, from the tobacco rhizosphere soils.
Results
The results showed that tobacco residue returning increased the diversity of microbial communities (bacteria and fungi) and changed the species composition. It further increased the relative abundance of beneficial microorganisms. After tobacco residue returning, the structure of the rhizosphere soil microbial community network was found more complex with strong interactions among microbial species. In addition, the keystone species of bacterial and fungal communities associated with tobacco rhizosphere soil were altered. This had exerted a driving force to the beneficial bacteria such as Sphiningomonas and Psathyrella to the keystone microorganisms which played important roles in microbial species interaction.
Conclusions
Tobacco residue return into soil showed significant effects on the microbial diversity, community composition, network structure, keystone microorganisms, and ecosystem functions of tobacco rhizosphere soils. This study provides a scientific basis for the improvement of tobacco field ecosystem functioning and ensuring soil health.
Similar content being viewed by others
Introduction
Tobacco-rice multiple cropping rotation is one of the main double-cropping systems in tobacco planting areas in Hunan, Jiangxi, and Guangdong and in other provinces of China (Hu et al. 2021). In this system, planting tobacco in spring and rice in autumn in the same farmland and year are practiced. The cropping system, thus mentioned, can improve the physicochemical properties of soil ensuring a full use of soil fertility. It also helps to reduce the occurrence of tobacco pests and diseases and the damage caused by weeds in the field. As a result, a good harvest of both grain and tobacco can be obtained (Zou et al. 2018). The cropping system also played a major role in stabilizing tobacco cultivating fields with improved yield and quality in the major tobacco farming areas of China (Zhang et al. 2015). A large number of tobacco residues are produced in the farming patterns of tobacco-rice multiple cropping rotation. Returning the residues (stem and root) into the field is a common agronomic practice. Previous studies indicated that returning tobacco residues to the field can improve soil organic matter and physical and chemical properties (Murage et al. 2007; Wang et al. 2008). However, systematic research to find the effects of tobacco residue returning into the tobacco-grown fields on the rhizosphere microbial community is still limited.
Soil microorganisms are one of the components of the agroecosystem functioning and contribute to the formation and development of soil, material recycling, fertility evolution, ecological functions, etc. (Singer 2008). The plant rhizosphere is considered to be the most active region in the soil and plays an important role in the growth, health, and productivity of plants (Luan et al. 2021). Plant rhizosphere microbial communities were found closely related to climate, soil types, agricultural management practices, and plant species (Berg et al. 2009; Garbeva et al. 2004; Jangid et al. 2008). And as a whole, soil microorganisms are involved in the decomposition of the plant residues, running nutrient cycles, and the corresponding responses of the soil microbial community to plant residue returning function precisely (Williams et al. 2006; Xu et al. 2020). Previous studies have shown that the return of crop residues into the soil ecosystems can elevate water retention of crop soil, improve soil structure, enhance soil fertility, increase soil microbial diversity, and change microbial community structure (Luan et al. 2021; Chen et al. 2021; Cui et al. 2021). However, the response of soil microbial communities to crop straw returning may be inconsistent with respect to different climates, regions, soil types, agricultural measures, and crops (Chen et al. 2015). At present, there is a dearth of knowledge on the systematic understanding of the effects of tobacco residue returning on the microbial diversity, community structure and composition, and network interactions among species of tobacco rhizosphere microbial communities.
The aims of this study are to (i) compare the bacterial and fungal diversity and community composition and structural differences between tobacco residue returned and control tobacco rhizosphere; (ii) reveal the effects of tobacco residue returning on the molecular ecological network and on the interactions between species through network analysis, and (iii) predict the differences of ecological function between tobacco residue returned and control tobacco rhizosphere. This study provided a scientific basis for improving the ecosystem multifunctionality of a tobacco field and ensuring soil health.
Materials and methods
Materials
The outdoor pot experiment was conducted in the Key Laboratory of Tobacco Science & Health of Hunan Agricultural University for three consecutive years (2018–2020). The tobacco (Nicotiana tabacum L., Family: Solanaceae) residues were composed of stems and roots and collected from the tobacco field located in Ningxiang County, Hunan Province, China. The experiment was carried out in plastic pots having individual size of 71.0 × 45.5 × 18.0 cm. The rice (Oryza sativa L., Family: Poaceae) variety used in the experiment was Xiangzaoxian No. 45 which was hybridized by the Agricultural Science Institute, Yiyang City, Hunan Province. The experiment was based on the crop rotation cycle with late rice cultivation after the harvest of flue-cured tobacco. The late rice variety, Xiangzaoxian No. 45 is a conventional medium maturity type early indica rice and widely cultivated in Hunan Province as a double-cropping early rice, having a whole growth period of about 106 days with a high yield and a good quality.
Experimental design
In July 2018, the stems and roots of the tobacco were cut into 5–10 cm pieces and dried. An amount of 25 kg of air-dried soil and the same amount of water were added into each plastic pot and mixed thoroughly with the designed amount of tobacco residues. Two treatments and 8 replicates for each treatment were designed, one named as PT (60 g tobacco stems + 40 g tobacco roots per pot, dry matter weight) and the other was a control CK (no tobacco residue returning). The late rice seeds were sown in the pots. In 2019, after the planting of tobacco in the 2018-potting soils, 60 g (dry weight) of tobacco stems + 40 g (dry weight) of tobacco roots was added to each pot, then rice was planted. In March 2020, tobacco was planted in the 2019-potting soils, for a total of two tobacco-rice cycles across 3 years.
Soil sampling
After 80 days of tobacco growth, the adhering rhizosphere soil samples after shaking off bulk soils were collected in PBS (0.1% Tween 80) with a brush. After removing the gravel and unwanted plant debris from the soil, the target sample was isolated and quickly placed in a labeled 50-mL sterilized centrifuge tube. The sample was frozen at − 20℃ in a refrigerator and preserved for future use.
DNA extraction and high-throughput sequencing
The total DNA was extracted using the FastDNA™ Spin kit (MP Biomedicals) following the instructions. The method of Zhang et al. (2017) was used to conduct PCR amplification and high-throughput sequencing of 16S rRNA. The V5-V6 region of 16S rRNA was amplified using primers 799F (5′-AACMGGATTAGATACCCKG-3′)/1115R (5′-AGGGTTGCGCTCGTTG-3′) (Hu et al. 2020), and the internal transcribed spacer 2 (ITS2) was amplified using primers 5.8F (5′-AACTTTYRRCAAYGGATCWCT-3′)/4R (5′-AGCCTCCGCTTATTGATATGCTTAART-3′) (Tan et al. 2021). The PCR reaction system was 50 µL, including 5 µL DNA template, 2.5 U Taq DNA polymerase (TaKaRa), 1 × Taq buffer, 75 µM dNTPs, and 0.3 µM primers (Gao et al. 2022; Jin et al. 2021). The PCR program for ITS2 was as follows: 94 °C for 1 min; followed by 35 cycles of (94 °C for 20 s, 57 °C for 25 s, and 68 °C for 45 s) and then 68 °C for 10 min. The PCR products were stored at 4 °C. The recovered product was qualified and quantitatively determined by a nanodrop spectrophotometer (Nano-100, Aosheng Instrument Co., Ltd.). Subsequently, the purified amplicons were pooled together and were used to prepare the sequencing library with VAHTS™ Nano DNA Library Prep Kit, subsequently sequenced on the Illumina HiSeq 2500 System.
Amplicon sequence processing and analysis
The raw sequences were quality screened and analyzed with the Quantitative Insights into Microbial Ecology (QIIME) pipeline (v1.9.1). After the barcode and primer sequences were removed, the opposite end sequences of the 16S rRNA gene were merged, and their quality was checked by Flash Program (Kong et al. 2011). The ITS2 sequence of forward and reverse primers were removed. Subsequently, the sequences were passed through the ITX program to remove the ITS flanking regions and non-fungal sequences. Next, sequences were clustered into operational taxonomic units (OTUs) using UPARSE with a sequence similarity threshold of 97%. At the end, OTU tables were created for downstream bioinformatic analysis.
Microbial network construction and analysis
The OTUs of microorganisms (fungi and bacteria) obtained by high-throughput sequencing were analyzed on the Molecular Ecological Network Analyses Pipeline (MENA) website (http://ieg4.rccc.ou.edu/mena). Based on random matrix theory (RMT) with threshold set, the data thus obtained was used to construct phylogenetic molecular ecological networks (pMEN) of tobacco rhizosphere microorganisms for the two treatments PT and CK, with network topology parameters generated. Microbial networks were visualized and analyzed by Gephi (0.9.2).
Data analysis
Two measurements of alpha-diversity were calculated to assess the biodiversity of tobacco rhizosphere microbial community. Richness was obtained by counting the number of species displayed in the OTU table. Shannon diversity index was calculated using the “vegan” package in R. The Alpha diversity indices (Richness and Shannon) were calculated using “alpha_diversity.py” from QIIME. The significant differences were checked using analysis of variance (ANOVA) and Student’s t test. And the differences for microbial community structures were investigated by detrended correspondence analysis (DCA).
Results
Effects of tobacco residue return on rhizosphere microbial diversity and community structure
The bacterial diversities of tobacco rhizospheres with control (CK) and tobacco residue (PT) treatment based on 16S rDNA sequencing are shown in Fig. 1a, b. According to the diversity indices Richness and Shannon (Fig. 1b), the bacterial diversity of tobacco rhizosphere was significantly different between CK and PT (P < 0.01), with significantly higher bacterial diversity in PT than that of CT. The fungal diversities of tobacco rhizospheres with CK and PT treatments based on ITS sequencing are shown in Fig. 1c, d. According to the diversity indices Richness and Shannon, the rhizosphere fungal community diversity was significantly different between CK and PT (P < 0.01), with significantly higher fungal diversity in the PT treatment. The results demonstrated that the returning of tobacco residues enhanced the diversity of both bacterial and fungal communities in the tobacco rhizosphere.
Microbial diversities of tobacco rhizospheres with the CK and PT treatments based on 16S and ITS sequencing (a bacterial Richness index, b bacterial Shannon index, c fungal Richness index, d fungal Shannon index). CK and PT represent the control treatment and tobacco residue treatment, respectively, similarly hereinafter. * < 0.05; ** < 0.01
The DCA was performed to test the differences for bacterial (Fig. 2a) and fungal (Fig. 2b) community structures between CK and PT treatments. Both the bacterial or fungal community structures of CK- and PT-treated rhizosphere soils were clearly separated. The results showed that the returning of tobacco residues altered the microbial (bacterial and fungal) community structure.
Effects of tobacco residue return on microbial community composition
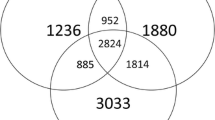
Based on 16S rRNA and ITS high-throughput sequencing results, Venn diagrams were made to investigate the shared and unique OTUs between CK- and PT-treated rhizosphere soils (Fig. 3). The results showed that PT-treated rhizosphere soils had higher number and percent of unique OTUs for both bacteria (Fig. 3a) and fungi (Fig. 3b), but lower percent of shared OTUs compared to CK treatments.
The microorganisms were identified and classified, and the detected OTUs for bacteria were classified into 8 phyla, and 15 genera and for fungi 6 phyla, and 18 genera with relative abundance > 1%. The phylum-level compositions of bacterial communities (relative abundance > 1%) in the CK and PT treatments are shown in Fig. 4a. The bacterial phyla with the highest relative abundance were Actinobacteria and Proteobacteria, followed by Firmicutes, Bacteroidetes, Acidobacteria, and Chloroflexi. The genus-level compositions of bacterial communities (relative abundance > 1%) in the CK- and PT-treated rhizosphere soils are displayed in Fig. 4b. The bacterial genus with the highest relative abundance was Arthrobacter, followed by Massilia, Sphingomonas, Gaiella, Terrabacte, and Nocardioides. The bacterial genera also included Rhodanobacter, Pseudomonas, Bradyrhizobium, Burkholderia, Falsibacillus, Conexibacter, and Clostridium sensu stricto. The phylum-level compositions of fungal communities (relative abundance > 1%) in the CK- and PT-treated rhizosphere soils are exhibited in Fig. 4c. The fungal phylum with the highest relative abundance was Ascomycota, followed by Zygomycota and Basidiomycota. The fungal phyla also included Chytridiomycota and Glomeromycota. The genus-level compositions of fungal communities (relative abundance > 1%) in the CK- and PT-treated rhizosphere soils are shown in Fig. 4d. The fungal genera with the highest relative abundance were Aleuria and Mortierella, followed by Cyberlindnera and Plectosphaerella. The fungal genera also included Entoloma, Podospora, Cryptococcus, Guehomyces, Gibberella, Rhodotorula, Monoblepharis, Conocybe, Fusarium, Paraphoma, Paraconiothyrium, and Mucor.
Comparison of microbial community compositions of tobacco rhizosphere between the CK- and PT-treated rhizosphere soils (a comparison of bacterial OTUs at the level of phylum with relative abundance ≥ 1%, b comparison of bacterial OTUs at the level of genus with relative abundance ≥ 1%, c comparison of fungal OTUs at the level of phylum with relative abundance ≥ 1%, d comparison of fungal OTUs at the level of genus with relative abundance ≥ 1%)
Further, the significant differences of microbial community compositions between CK- and PT-treated rhizosphere soils at the phylum level were investigated, and the results are listed in Table 1. At the bacterial phylum level, the relative abundance of Actinobacteria in PT-treated rhizosphere soil was significantly increased compared to CK (P < 0.01), while the relative abundances of Proteobacteria, Firmicutes, and Chloroflexi were significantly decreased (P < 0.01 or P < 0.05), and no significant differences were seen for the other bacterial phyla. At the fungal phylum level, the relative abundance of Chytridiomycota in PT-treated rhizosphere soil was significantly increased compared to CK rhizosphere soil (P < 0.05), while the relative abundance of Zygomycota was significantly lower (P < 0.05). And no significant differences were seen for the other fungal phyla. The significant differences of microbial community compositions between CK- and PT-treated rhizosphere soils at the genus level are shown in Table 2. At the bacterial genus level, the relative abundance of Arthrobacter in PT-treated rhizosphere soil was significantly higher than that of CK (P < 0.001), while the relative abundances of Sphingomonas, Terrabacter, Conexibacter, Falsibacillus, and Clostridium sensu stricto were significantly lower (P < 0.01 or P < 0.05). At the fungal genus level, the relative abundances of Mortierella, Gibberella, Conocybe, Paraphoma, and Mucor in PT-treated rhizosphere soil were significantly higher than those of CK (P < 0.01 or P < 0.05), while the relative abundances of Aleuria, Plectosphaerella, Entoloma, Guehomyces, and Fusarium were significantly lower (P < 0.01 or P < 0.05). Therefore, the tobacco residue returning altered the bacterial and fungal community compositions of rhizosphere soils.
Effects of tobacco residue return on bacterial community network structure and keystone species
MENA was carried out to explore the effects of tobacco residue returning on the species interactions in bacterial communities. In the bacterial community network of tobacco rhizosphere, compared with CK rhizosphere soil, the bacterial network of PT-treated rhizosphere soil showed more nodes and links, exhibiting rather a complex structure. The topological characteristics are listed in Table 3. Regarding network nodes, CK rhizosphere soil had 107 and PT-treated rhizosphere soil had 217 and thus for the number of links, and CK- and PT-treated rhizosphere soils had 103 and 257, respectively. On the other hand, the number of modules, CK- and PT-treated rhizosphere soils had 34 and 41, respectively. In addition, the average clustering coefficient (avgCC) of CK- and PT-treated rhizosphere soils in the empirical network (0.46 and 0.34, respectively) were higher than the corresponding avgCC in the random network (0.030 and 0.051, respectively), and the modularity (M) values of CK- and PT-treated rhizosphere soils in the empirical network (0.83 and 0.86, respectively) were higher than the corresponding M values in the random network (0.754 and 0.716, respectively). The results indicated that the constructed networks had small-world network characteristics (Watts and Strogatz 1998) and modular topology features (Newman 2016). The constructed networks were visualized, with the bacterial networks of CK- and PT-treated rhizosphere soils shown in Fig. 5a, b. Overall, there were significant differences in the bacterial community network structures of tobacco rhizosphere between the CK and PT treatments.
Furthermore, a Zi-Pi plot was made to demonstrate the distribution of OTUs playing typical topological roles in tobacco rhizosphere bacterial network structures of the CK and PT treatments (Fig. 6). Based on the thresholds set for Zi = 2.5 and Pi = 0.62, all nodes in the network were distributed in the four subcategories: peripherals, connectors, module hubs, and network hubs. The latter three of which were recognized as keystone microbial species that played important roles in ecological networks (Zhou et al. 2011). It can be seen from the plot that no keystone species were shown in the network structure of CK rhizosphere soil. Whereas in the network structure of PT-treated rhizosphere soil, OTU_54 (incomplete taxonomic information) and OTU_65 (genus: Sphingonas) were distributed in the module hubs. It can be seen that the tobacco residue returning resulted in the emergence of keystone species in the bacterial community network structure.
Effects of tobacco residue return on fungal community network structure and keystone species
MENA was again conducted to investigate the effects of tobacco residue return on the species interactions in fungal communities. In the fungal community network of tobacco rhizosphere, the fungal network structure of PT-treated rhizosphere soil exhibited more network nodes and links. In addition, compared to CK, the fungal network also showed a more complex structure in PT rhizosphere soil. As shown in Table 4, the network nodes in CK- and PT-treated rhizosphere soil were 174 and 179, respectively. Regarding the number of links, CK- and PT-treated rhizosphere soil had 232 and 367, respectively. But the number of modules, in CK- and PT-treated rhizosphere soil were 21 and 15, respectively. In addition, the avgCC values of CK- and PT-treated rhizosphere soils in the empirical network (0.28 and 0.32, respectively) were higher than the corresponding avgCC values in the random network (0.034 and 0.060, respectively). And the M values of CK- and PT-treated rhizosphere soils in the empirical network (0.67 and 0.81, respectively) were higher than the corresponding M values in the random network (0.52 and 0.67, respectively). The results indicated that the networks we constructed had small-world network characteristics (Watts and Strogatz 1998) and modular topology features (Newman 2016). The constructed networks were visualized, with the fungal networks of CK- and PT-treated rhizosphere soils shown in Fig. 7a, b. Overall, there were significant differences in the fungal community network structure of tobacco rhizosphere soils between the CK and PT treatments.
Furthermore, a Zi-Pi plot was also made to demonstrate the distribution of OTUs playing typical topological roles in tobacco rhizosphere fungal network structures of the CK and PT treatments (Fig. 8). The classification information of the keystone OTUs in the Zi-Pi plot is shown in Tables 4 and 5. In the CK network structure, OTU_13 (genus: Cyberlindnera), OTU_347 (genus: Ochroconis), OTU_11714 (genus: Monoblepharis), OTU_15497 (genus: Talaromyces), and OTU_44 (genus: Humicola) were the keystone fungi. While in the PT treatment network structure, we found that OTU_18 (genus: Cyberlindnera), OTU_350 (class: Agaricomycetes), OTU_419 (genus: Psathyrella), and OTU_156 (genus: Mortierella) were the keystone fungi. It can be seen that the tobacco residue returning changed the keystone species in the fungal community network structure.
Effects of tobacco residue return on the ecological function of microbial communities
Functional Annotation of Prokaryotic Taxa (FAPROTAX) was used to predict the bacterial community functions of tobacco rhizosphere in CK and PT treatments (Louca et al. 2016), resulting a total of 58 functional categories. The ecological functions of bacterial communities with relative abundances > 1% are shown in Fig. 9a, including fermentation, aerobic chemoheterotrophy, aromatic compound degradation, ureolysis, and chemoheterotrophy. Interestingly, significant differences were shown in aerobic chemoheterotrophy, chemoheterotrophic, and aromatic compound degradation functions in the rhizosphere bacterial communities between CK- and PT-treated rhizosphere soils, and their relative abundances in PT-treated rhizosphere soil were significantly higher than those of CK (P < 0.05).
FUNGuild (“Fungi” + “Functional” + “Guild”) (Nguyen et al. 2016) was used to perform functional prediction of tobacco rhizosphere fungal communities of CK and PT treatments. In this study, a total of 4 fungal community functions with relative abundance > 1% were identified (Fig. 9b). They were pathotroph, saprotroph, symbiotroph, and others. Interestingly, there were significant differences in pathotroph and saprotroph functions of rhizosphere fungal communities between CK and PT treatments. The pathotroph function in PT-treated rhizosphere soil was significantly lower than that of CK (P < 0.05), whereas the saprotroph function was significantly higher than that of CK (P < 0.01).
Discussion
Soil is one of the most complex ecosystems of our biosphere where a rich variety of microbial communities does exist and can act as a microbial reservoir (Mason-Jones et al. 2021). Soil microbiome is a collective term used to express the existence of its massive microorganisms such as bacteria, archaea, fungi, viruses, protists, and other microbial groups (Muller et al. 2016). They play an important role in maintaining soil health, participating in the cycling of soil organic matter, nitrogen, phosphorus, and potassium, and engaging in biodegradation and biocomposting processes, as well as improving soil fertility (Kong 2020; Jan et al. 2020). Soil microbial diversity, community composition and structure, and community molecular ecological network are the important indexes reflecting the status of soil nutrient cycling and farmland habitat stability (Wang et al. 2018). In recent years, with the development of microbial high-throughput sequencing technology and bioinformatics, the study of rhizosphere soil microbiome has entered into an unprecedented era of development (Gao et al. 2020). When the tobacco residues were returned into the field, with the increase of the returning years, a series of physicochemical changes occurred in the residues under the conditions of soil water and heat and had affected the microbial diversity and community composition in the soil (Chen et al. 2021; Song et al. 2021). Therefore, it is of great significance to systematically study the effects of tobacco residue returning on the microbial community of tobacco rhizosphere soil.
In this study, the effect of tobacco residue returning on rhizosphere soil microbial community was comprehensively analyzed using high-throughput sequencing and bioinformatics. The results showed that the treatment of tobacco residue returning could increase rhizosphere soil microbial diversity to a certain extent. As it has been indicated by the Richness and Shannon indices of bacterial and fungal communities. Soil microbial diversity is of great positive significance in ensuring the quality of soil, maintaining soil ecosystem function, disease resistance, and promoting healthy growth of crops (Ananthacumaraswamy et al. 2011). Tobacco residue returning treatment changed the composition of microbial community. At the bacterial phylum level, the relative abundance of Actinobacteria in PT-treated rhizosphere soil was significantly higher than that in CK rhizosphere soil (P < 0.01). Many members of phylum Actinobacteria were thought to have the potential to degrade lignocellulose, which may contribute to the degradation of tobacco residues (Rong et al. 2021). At the fungal phylum level, on the one hand, the relative abundance of Chytridiomycota in PT-treated rhizosphere soil was significantly increased compared to that in CK rhizosphere soil (P < 0.05). While on the other hand, the relative abundance of Zygomycota was significantly reduced (P < 0.05). Most members of the phylum Chytridiomycota favored to live in dry or moist soils, while members of phylum Zygomycota might be phytopathogenic (Voigt et al. 2014). At the bacterial genus level, the relative abundance of Arthrobacter in PT-treated rhizosphere soil was significantly higher than that in CK rhizosphere soil (P < 0.001). Many members of this genus were reported with good bioremediation effects on soils (Li et al. 2017). At the fungal genus level, the relative abundances of Mortierella and Gibberella in PT-treated rhizosphere soil were significantly higher than those in CK rhizosphere soil (P < 0.01 or P < 0.05). The secondary metabolites of these fungi were reported with antibacterial and bacteriostatic abilities. Whereas the relative abundances of Aleuria, Plectosphaerella, Entoloma, Guehomyces, and Fusarium were significantly lower than those in CK rhizosphere soil (P < 0.01 or P < 0.05), among which Fusarium was a soil-borne root rot pathogen of tobacco (Yang et al. 2020). It can be seen that the tobacco residue returning treatment increased the microbial diversity, changed the composition of the microbial (bacteria and fungi) community, and boosted the abundance of beneficial microorganisms.
In the plant rhizosphere soil micro-ecosystem, the interactions between microbial species play a crucial role in the functional diversification and stability of microbial communities (Lurgi et al. 2014). In recent years, molecular ecological network analysis has been widely used to explore the interactions between microorganisms in various micro-ecological environments and to explore the keystone species in the network (de Menezes et al. 2015). The results of the present study demonstrated that the soil bacterial and fungal community network structures of tobacco residue returning treatment exhibited more topology-characterized network nodes, links, and complexities than the control. Previous studies have shown that the interactions among microorganisms within the microbial community lead to the complexity of the community structure and maintained the ecosystem stability (Zhang et al. 2014). In the tobacco rhizosphere of PT treatment, more connections between OTUs were exhibited, indicating more interactions between species in the microbial community of PT-treated rhizosphere soil. This result also showed that the exchange of information, energy, and nutrients among different microorganisms within PT-treated rhizosphere soil may be more abundant and more diverse and may better cope with different biotic and abiotic stresses and better maintain the soil micro-ecosystem functions (Chen et al. 2018).
Additionally, the keystone species are thought to play an essential role in the microbial community network. Zi-Pi plots showed that OTU_54 (incomplete taxonomic information) and OTU_65 (genus: Sphiningomonas) of bacterial community within PT-treated rhizosphere soil were located in the module hubs, whereas no keystone species were found in CK rhizosphere soil network. The genus Sphingomonas has a variety of functions from remediating environmental pollution to producing highly beneficial plant hormones and promoting plant healthy growth under adverse conditions such as drought, salinity, and heavy metals in agricultural soils and are well recognized as soil beneficial bacterium (Asaf et al. 2020). In the fungal network structure of PT-treated rhizosphere soil, it was revealed that OTU_18 (genus: Cyberlindnera), OTU_350 (class: Agaricomycetes), OTU_419 (genus: Psathyrella), and OTU_156 (genus: Mortierella) were the keystone fungi. And many secondary metabolites of these fungi (such as Psathyrella and Mortierella) have been reported with antibacterial and bacteriostatic abilities (Li et al. 2018; Gross et al. 2018). They are potential strains for the biological control of plant diseases.
Based on FAPROTAX function prediction (Fig. 9), the relative abundances of aerobic heterotrophic, chemoheterotrophic, and aromatic compound degradation functions in the bacterial community of tobacco rhizosphere with PT treatment were higher than those in CK. Chemoheterotrophic bacteria are generally considered to play an important role in the cycling of organic matter in all ecosystems (Liu et al. 2022). Furthermore, chemoheterotrophy was the primary pathway of carbon cycling in microbial communities (McKinley and Wetzel 1979). The degradation function of aromatic compounds had been proven to be beneficial to the degradation and restoration of returned residues, environmental pollutants, and pesticide residues (Seo et al. 2009). Based on the FUNguide function prediction, the pathogenic function of the rhizosphere fungal community in PT-treated rhizosphere soil was significantly lower than that in CK (P < 0.05), whereas the saprotroph function was significantly higher than that in CK (P < 0.01). The results were consistent with the Zi-Pi plot result that PT treatment altered the keystone microbes, with beneficial microbes playing an important role in the fungal community and rendering rhizosphere pathogenic fungi recessive. It is speculated that these potential functions may serve as the basis for future regulation of rhizosphere soil microbial communities and an improvement of ecosystem functions.
Conclusion
In this study, tobacco residue return to the soil showed significant impacts on the microbial diversity, community composition, network structure, keystone microorganisms, and ecosystem functions in tobacco rhizosphere. According to the Shannon and Richness diversity indices, PT-treated rhizosphere soil had higher levels of bacterial and fungal diversities than CK rhizosphere soil. The DCA results demonstrated a clear separation between CK- and PT-treated rhizosphere soils regarding bacterial and fungal community structures. Molecular ecological network analysis showed more complex network structures of bacterial and fungal communities in PT-treated rhizosphere soil. And PT treatment changed the keystone microorganisms of the bacterial and fungal communities in tobacco rhizosphere. In addition, PT treatment demonstrated better microbial ecological functions by function predictions.
Availability of data and materials
16S rRNA gene and ITS sequencing data of all samples were submitted to the China National Microbiology Data Center (https://nmdc.cn) under the accession SUB1661165384987.
References
Ananthacumaraswamy S, Hettiarachchi LSK, Dissanayake SM (2011) Soil and foliar sulfur status in some tea plantations of Sri Lanka. Commun Soil Sci Plant Anal 34(11–12):1481–1497
Asaf S, Numan M, Khan AL, Al-Harrasi A (2020) Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit Rev Biotechnol 40(2):138–152
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68(1):1–13
Chen LJ, Sun B, Jin C, Jiang YJ, Chen L (2015) Effects of organic fertilizer and biochar with equal carbon input on microbial diversity and soil respiration in red soil. Soil. 47(2):340–348
Chen S, Qi GF, Luo T, Zhang HC, Jiang QK, Wang R, Zhao XY (2018) Continuous-cropping tobacco caused variance of chemical properties and struproperties and structure of bacterial network. Land Degrad 29:4106–4120
Chen H, Rao JX, Sun QY (2021) Comprehensive effect evaluation of straw returning methods on soil physicochemical properties. J Anhui Agric Univ 48(4):661–667
Cui HZ, An YY, Li GW, Fu YZ, Ma K (2021) Effects of corn stalks returning on soil microbial community composition. Agric Sci Res 42(2):10–16
De Menezes AB, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH (2015) Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ Microbiol 17(8):2677–2689
Gao GF, Chu HY (2020) Microbiome technologies and methods and their applications. Chin J Plant Ecolog 44(4):395–408
Gao HJ, Li Q, Zhu M, Peng C, Zhang XZ, Jiao YF, Gao JC, Zhu P (2022) Effects of different crop rotation and straw returning methods on bacterial community structure in black soil. Journal of Jilin Agricultural University 44(3):336–344.
Garbeva P, Veen JAv, Elsas JDv (2004) Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Ann Rev Phytopathol 42(42):243–270
Gross S, Kunz L, Müller DC, Santos Kron A (2018) Characterization of antagonistic yeasts for biocontrol applications on apples or in soil by quantitative analyses of synthetic yeast communities. Yeast (Chichester, England) 35(10):559–566
Hu QL, Tan L, Gu SS, Xiao YS, Xiong XY, Zeng WA, Feng K, Wei Z, Deng Y (2020) Network analysis infers the wilt pathogen invasion associated with non-detrimental bacteria. NPJ Biofilms Microbi 6(1):e8
Hu R, Liu YJ, Chen T, Zheng Z, Peng G, Zou Y, Tang CG, Shan XH, Zhou QM, Li J (2021) Responses of soil aggregates, organic carbon, and crop yield to short-term intermittent deep tillage in Southern China. J Clean Prod 298:126767
Jan U, Rui F, Masood J, Chun SC (2020) Characterization of soil microorganism from humus and indigenous microorganism amendments. Mycobiology 48(5):392–398
Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, Jenkins MB, Endale DM, Coleman DC, Whitman WB (2008) Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem 40(11):2843–2853
Jin YT, Li XF, Cai Y, Hu HX, Liu YF, Fu SW, Zhang BR (2021) Effects of straw returning and chemical fertilizer application on enzyme activities and microbial community structure of rice-oilseed rotation soil. Environ Sci: 1–16.
Kong D (2020) Effects of straw returning and fertilization on soil carbon, nitrogen and microbial communities in wheat-soybean rotation soil. Northwest A&F University. Chinese doctoral thesis. https://doi.org/10.27409/d.cnki.gxbnu.2020.001483
Kong Y (2011) Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98:152–153
Li J, Constantine U, Leng Y, Li SW, Chen XM (2017) Research progress on the treatment of organic matter and heavy metal pollutants by Arthrobacter. Environ Sci Technol 40(10):89–97
Li F, Chen L, Redmile-Gordon M, Zhang J, Zhang C, Ning Q, Li W (2018) Mortierella elongata’s roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad Dev 29(6):1642–1651
Liu HW, Du XF, Li YB, Han X, Li B, Zhang XK, Li Q, Liang WJ (2022) Organic substitutions improve soil quality and maize yield through increasing soil microbial diversity. J Clean Prod 347:131323
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277
Luan L, Zheng J, Cheng MH, Hu KJ, Kong PJ, Jiang YJ, Sun B (2021) Effects of different straw returning methods on bacterial diversity and community structure in dryland red soil. Soil 53(5):991–997
Lurgi M, Galiana N, López BC, Joppa LN, Montoya JM (2014) Network complexity and species traits mediate the effects of biological invasions on dynamic food webs. Frontiers Ecol Evol 2:1–11
Mason-Jones K, Robinson SL, Veen GF, Manzoni S, van der Putten WH (2021) Microbial storage and its implications for soil ecology. ISME J 16(3):617629
McKinley KR, Wetzel RG (1979) Photolithotrophy, photoheterotrophy, and chemoheterotrophy: patterns of resource utilization on an annual and a diurnal basis within a pelagic microbial community. Microb Ecol 5(1):1–15
Muller DB, Vogel C, Bai Y, Vorholt JA (2016) The plant microbiota: systems-level insights and perspectives. In: Bonini NM, editor. Annual Review of Genetics Vol. 50. Annual Reviews, Palo Alto, pp. 211–234.
Murage EW, Voroney PR, Kay BD, Deen B, Beayaert RP (2007) Dynamics and turnover of soil organic matter as affected by tillage. Soil Sci Soc Am J 71(4):1363–1370
Newman MEJ (2016) Modularity and community structure in networks. Pro Natl Acad Sci USA 103(23):8577–8582
Nguyen NH, Song ZW, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Rong LL, Zhao LF, Zhao LC, Cheng ZP, Yao YM, Yuan CL, Wang L, Sun HW (2021) LDPE microplastics affect soil microbial communities and nitrogen cycling. Sci TotalL Environ 773:145640
Seo JS, Keum YS, QiX Li (2009) Bacterial degradation of aromatic compounds. Int J Environ Res Public Health 6(1):278–309
Singer A (2008) Soil horizon designations in the WRB soil classification system. Encyclopedia of Soil Science. pp 668–670
Song SL, Wu H, Huang PW, Sun K, Zhang ZH, Zhang Y, Dai CC (2021) Effects of straw returning for soil improvement and fertilization matrix and the application of compound inoculants on soil ecology. J Ecol 41(11):4562–4576
Tan L, Zeng WA, Xiao YS, Li PF, Gu SS, Wu SL, Zhai ZG, Feng K, Deng Y, Hu QL (2021) Fungi-bacteria associations in wilt diseased rhizosphere and endosphere by interdomain ecological network analysis. Front Microbiol 12:E722626
Voigt K, Kirk PM (2014) FUNGI-Classification of Zygomycetes. Encyclopedia of Food Microbiology, 2nd edn. SAS, USA
Wang P, Zeng LL, Wang FP, Zhang XM, Bi H, Li ZH (2008) Effects of straw returning on nitrogen accumulation, distribution and utilization in flue-cured tobacco. Soil Fertilizer Sci China 4:43–46
Wang HY, Xu MG, Ma X, Duan YH (2018) Research progress of farmland soil microorganisms and ammonia oxidizing bacteria in China under long-term fertilization. China Soil Fertilizer. (02):1–12.
Watts DJ, Strogatz SH (1998) Collective dynamics of ‘small-world’ networks. Nature 393:440–442
Williams MA, Myrold DD, Bottomley PJ (2006) Carbon flow from 13C-labeled straw and root residues into the phospholipid fatty acids of a soil microbial community under field conditions. Soil Biol Biochem 38:759–768
Xu YD, Sun LJ, Lal R, Bol R, Wang Y, Gao XD, Ding F, Liang SW, Li SY, Wang JK (2020) Microbial assimilation dynamics differs but total mineralization from added root and shoot residues is similar in agricultural Alfisols. Soil Biol Biochem 148:e107901
Yang M, Cao JD, Zheng YX, Wang JM, Zhou HF, He MD, Duan J, Tong WJ, Deng XP, Chen XL, Yan D, Cai XJ, Zhong Y, Huang FY, He YS, Yu L (2020) First report of Fusarium root rot of tobacco caused by Fusarium solani in Lincang. China Plant Dis 104(5):1541–1542
Zhang R, Vivanco JM (2017) Shen Q (2017) The unseen rhizosphere root-soil-microbe interactions for crop production. Curr Opin Microbiol 37:8–14
Zhang Y, Cong J, Lu H, Yang C, Yang Y, Zhou J, Li DQ (2014) An integrated study to analyze soil microbial community structure and metabolic potential in two forest types. PLoS ONE 9:e93773
Zhang Z, Cui B, Li Y, Liu G, Xiao H, Liao Y, Li Y, Zhang Y (2015) Effects of tobacco-rice rotation on rice planthoppers Sogatella furcifera (Horvath) and Nilaparvata lugens (Stal) (Homoptera: Delphacidae) in China. Plant Soi 392:333–344
Zhou J, Deng Y, Luo F, He Z, Yang Y (2011) Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio. 2(4):e00122-11
Zou C, Li Y, Huang W, Zhao G, Pu G, Su J, Coyne MS, Chen Y, Wang L, He X, Jin Y (2018) Rotation and manure amendment increase soil macro-aggregates and associated carbon and nitrogen stocks in flue-cured tobacco production. Geoderma 325:49–58
Acknowledgements
This research was supported by the Foundation for Tobacco Science of Changsha Tobacco Company of Hunan Province (No.2018-001) and Wenshan Tobacco Company of Yunnan Province (2021530000241033) of China. This paper was jointly supported by Hunan Agricultural University and Changsha Tobacco Company of Hunan. We thank Dr. James Walter Voordeckers for carefully editing the grammar of the manuscript and for some valuable suggestions for this paper.
Funding
This research was supported by the Foundation for Tobacco Science of Changsha Tobacco Company (No.2018–01) and Zhuzhou Tobacco Company (No.22–004) of Hunan Province in China.
Author information
Authors and Affiliations
Contributions
Jin Chen carried out the experiments, formal analysis, and wrote the article. Weiai Zeng contributed to the methodology and wrote the article. Yang Zhang carried out the experiments. Jie Huang carried out the experiments and formal analysis. Zhifeng Chen carried out the experiments. Jiguang He carried out the experiments. Xiaohua Deng contributed to the methodology and formal analysis and wrote the article. Qiulong Hu carried out the experiments. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study did not violate ethics, and all participants agreed to publish the paper.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, J., He, J., Zhang, Y. et al. Effects of tobacco plant residue return on rhizosphere soil microbial community. Ann Microbiol 72, 42 (2022). https://doi.org/10.1186/s13213-022-01699-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13213-022-01699-z