Abstract
Purpose
Data support and theoretical basis for bioremediation and treatment of petroleum-contaminated soils in the Loess hills of Yan’an, northern Shaanxi.
Methods
The evolutionary characteristics of soil microbial diversity and community structure under different levels of oil pollution were studied by field sampling, indoor simulation experiments, and analyzed through assays, using the mine soils from Yan’an, Shaanxi Province, as the research object.
Results
Compared with clean soil, the microbial species in contaminated soil were significantly reduced, the dominant flora changed, and the flora capable of degrading petroleum pollutants increased significantly. The soil microbial diversity and community structure differed, although not significantly, between different pollution levels, but significantly from clean soil. In the uncontaminated soil (CK), the dominant soil microbial genera were mainly Pantoea, Sphingomonas, Thiothrix, and Nocardioides. The abundance of Pseudomonas, Pedobacter, Massilia, Nocardioides, and Acinetobacter in the soil increased after oil contamination, while Thiothrix, Sphingomonas, and Gemmatimonas decreased significantly.
Conclusions
After the soil was contaminated with petroleum, the microbial species in the soil decreased significantly, the dominant genera in the soil changed, and the relative abundance of bacteria groups capable of degrading petroleum pollutants increased. The genera that can degrade petroleum pollutants in the petroleum-contaminated soil in the study area mainly include Pseudomonas, Acinetobacter, Pedobacter, Acinetobacter, and Nocardioides, which provide a scientific basis for exploring It provides a scientific basis for exploring remediation methods suitable for petroleum-contaminated soil in this region.
Similar content being viewed by others
Introduction
The loess hills area in northern Shaanxi, China, is an important oil and gas reserve and development area. Oil well leaks, crude oil pipeline, and storage tank leaks during the development of oil and gas fields cause petroleum-based products to fall on the ground soil, causing serious damage to the local ecological environment, in turn, affecting the ecological balance and leads to gradual environmental degradation (Zhen et al. 2015; Wang et al. 2017). Petroleum hydrocarbons are hard-to-degrade organic pollutants that can seriously affect soil productivity and cause different degrees of harm to microorganisms (Mambwe et al. 2021), plants, and animals in the ecosphere around the pollution source (Wang et al. 2016). The entry of petroleum-based substances into the soil has caused a dramatic increase in soil organic carbon content, resulting in an imbalance between soil C content and N content (He et al. 2021; Zhang et al. 2018; Wen et al. 2017). This imbalance destroys the habitat for soil microbial life and changes the number and diversity of soil microbial populations (Zheng et al. 2021; Dong 2020). Microorganisms play a very important role in maintaining soil ecosystem functions; they can effectively decompose plant and animal residues; promote soil C, N, S, P, and other nutrient cycles (Pan et al. 2012; Li et al. 2017; Mu 1994); and regulate soil material and energy cycle (Iyobosa et al. 2021; Kong et al. 2021; Wang et al. 2021). Therefore, it was important to investigate the current status of petroleum-contaminated soil in northern Shaanxi and analyze the effects of petroleum pollution on the diversity of soil microbial populations to explore remediation methods suitable for petroleum-contaminated soil in the region (Ajona and Vasanthi 2021; Fan et al. 2015).
The current research on microorganisms of petroleum-contaminated soil has mainly focused on the remediation effect of soil microorganisms (screening of efficient degradation bacteria, degradation characteristics, etc.) (Ueno et al. 2010), while the structural diversity of indigenous microbial communities of petroleum-contaminated soil in Loess hilly areas has been less studied (Margesin et al. 2007). In this study, we investigated the soil microbial diversity under the effects of different levels of oil pollution and analyzed the response of soil microbial community structure to oil pollution through field sampling (Yang et al. 2013), indoor simulation tests, detection, and analysis, to provide a scientific basis for bioremediation and treatment of oil-contaminated soil in the Loess hills of northern Shaanxi Province. This study aims to provide a scientific basis for the bioremediation and treatment of oil-contaminated soil in the Loess hills (Wang et al. 2020; Li et al. 2021).
Materials and methods
Sample collection
The test soil for this study was collected from a typical oil well area in Baota District, Yan’an City, Shaanxi Province, China. The soil samples were collected in the 0-30 cm range by the S-spotting method in the vicinity of oil wells, selected from areas not contaminated with oil, following the principles of random, equal, and multi-point mixing. The samples were then passed through a 2-mm sieve to remove impurities and preserved at low temperature for indoor simulation tests (Li et al. 2009).
Indoor simulation test
Crude oil was weighed according to the ratio of oil content of 15g/kg (C15), 30g/kg (C30), and 45g/kg (C45); added to clean soil; and mixed thoroughly to prepare different oil contamination gradient soil (Li et al. 2011). After 1 week of static aging, indoor simulated incubation tests were carried out, i.e., incubation under constant conditions (ambient temperature controlled at 25±2°C and soil moisture content maintained at 15%) for 21 days, with three replicates for each treatment (Table 1). After incubation, soil samples were collected from four different locations in each container using the multi-point mixed soil sample collection method and then mixed thoroughly to form a mixed sample of about 20 g for determining the microbiological properties of the soil (Liu et al. 2012, 2020).
Testing indicators and measurement methods
Soil microbial diversity was determined by DNA extraction and high-throughput sequencing, and the genomic DNA was extracted by CTAB or SDS method, followed by agarose gel electrophoresis to check the purity and concentration of DNA. The PCR was performed using specific primers with Barcode, Phusion High-Fidelity PCR Master Mix with GC Buffer from New England Biolabs, and high efficiency high fidelity enzymes to ensure amplification efficiency and accuracy. The primers corresponding to the region for the identification of bacterial diversity were 16S V4 region primers (515F and 806R) (Zhang and Xu 2022; Sun et al. 2018).
The PCR products were detected by electrophoresis using a 2% agarose gel; the PCR products that passed the test were purified by magnetic beads, quantified by enzyme labeling, mixed in equal amounts according to the concentration of PCR products, mixed thoroughly, and then detected by electrophoresis using a 2% agarose gel, and the target bands were recovered using a gel recovery kit provided by Qiagen (Fig. 1). The library was constructed using TruSeq DNA PCR-Free Sample Preparation Kit. The constructed library was quantified by Qubit and Q-PCR, and after the library was qualified, the library was sequenced using NovaSeq6000 (Lusk et al. 2021).
Statistical analysis of data
According to the Barcode sequence and PCR amplification primer sequence to split each sample data from the downstream data, truncated the Barcode and primer sequence and used FLASH (V1.2.7) to splice the reads of each sample (Liu and Peng 2001), the spliced sequence was the original Tags data (Raw Tags); the spliced Raw Tags, need to go through strictly. The spliced Raw Tags need to be filtered strictly to get the high-quality Tags data (Clean Tags). Referring to the quality control process of Tags in Qiime (V1.9.1), the following operations were performed: (a) Tags interception: Raw Tags were truncated from the first low-quality site where the number of consecutive low-quality bases (default quality threshold <=19) reached a set length (default length value is 3) (Ali et al. 2020); (b) Tags length filtering: Tags were intercepted to obtain the Tags. The Tags obtained after the above processing need to be processed to remove the chimeric sequences, and the Tags sequences were compared with the species annotation database to detect the chimeric sequences, and finally removed the chimeric sequences to obtain the final effective data (Effective Tags) (Wagh and Tiwari 2020).
Results and discussion
Dilution curve
For the dilution curve, a certain number of sequences were randomly selected from the sample, and the Alpha diversity index of these sequences corresponding to the sample was counted. The horizontal coordinate represents the number of sequencing strips randomly selected from the sample, and the vertical coordinate represents the number of operational taxonomic units (OTUs) that can be constructed based on this number of sequencing strips to plot the curve, and the adequacy of the current sequencing data volume is judged according to whether the curve reaches a flatness (Ma et al. 2020). According to Fig. 2, the microbial OTU in petroleum-contaminated soils was significantly reduced compared to the uncontaminated controls, i.e., the overall number of microbial species decreased. The number of soil microbial OTUs at different oil contamination concentrations was in the order of C15> C45> C30, where the difference between C30 and C45 was small, indicating that the number of soil microbial species first decreased and then leveled off as the contamination level increased (Chen et al. 2020).
Microbial alpha (α)-diversity—analysis for variability between groups
Alpha diversity (α-diversity) refers to the species diversity within a community or habitat, mainly focusing on the species diversity within the community. The common method is to calculate Chao, Shannon, Simpson, and coverage4 indexes based on OTU results for biodiversity analysis (Liu et al. 2021). In this study, the Shannon and Simpson indices were selected to characterize the α-diversity of soil contaminated with different additives and to examine the differences between treatments, the inter-group difference test of indices was used. The results showed that the microbial α-diversity of petroleum-contaminated soils (C15, C30, C45) was significantly lower than the control group with no petroleum contamination, but the differences in soil microbial α-diversity between varying concentrations of contamination did not reach a statistically significant level (Fig. 3).
Microbial species composition analysis through Venn diagrams
Venn diagrams were obtained by comparing OTUs between samples or between subgroups, which can visualize the similarity and overlap of OTU composition of environmental samples (Wang et al. 2019). The results showed that each soil sample produced a total of 14,090 OTUs, of which 1791 were shared, accounting for 12.71% of the total OTUs (Fig. 4). the number of OTUs in the C15, C30, and C45 treatments were 3719, 3194, and 3345, respectively, which were lower than that of CK (3832). The number of shared OUTs between contaminated and clean soils showed a decreasing trend as the oil contamination gradient increased. The highest overlap between different pollution gradients was found in the C15 and C45 treatments, which had a high similarity of bacterial community structure. In addition, the different oil contamination treatments did not significantly affect the number of soil-specific OTUs.
Microbial β-diversity analysis for variability between groups
To study the similarity or difference relationship of different sample community structures, cluster analysis was performed on the sample community distance matrix to build a sample hierarchical clustering tree. This method effectively identifies the “major” elements and structures in the data, removes noise and redundancy, reduces the dimensionality of the original complex data, and reveals the simple structure hidden behind the complex data (Zhang et al. 2017).
Figure 5 shows that the different shape legends in the figure represent the control and three soil samples with different concentrations of oil contaminants. The differences in the microbial community composition of CK, C15, C30, and C45 were found by principal component analysis. The microbial community composition of the contaminated soil differed, although not significantly, between the different contamination treatments and differed significantly From CK. the PC1 and PC2 axes explain 20.03% and 9.23% of the results, respectively.
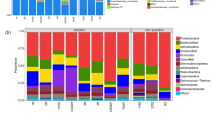
Differential analysis of microbial community composition
The histogram presents the community composition and species abundance at different taxonomic levels (Zhu et al. 2020). In this study, community composition and species abundance analysis were conducted at the genus level. In the absence of oil contamination (CK), the dominant genera included Pantoea, Streptomyces, Alkanindiges, and Massilia. After oil contamination, the dominant genera of the microbial community changed significantly, with Pantoea being dominant. After D2 treatment, the abundance of Streptomyces increased, and the difference between C30 and C45 decreased (Fig. 6). In addition, the abundance of Nocardioides and Acinetobacter increased with increasing oil pollution concentration, while Thiothrix, Sphinggomonas, and Gemmatimonas decreased significantly.
Conclusions
In this study, we used high-throughput sequencing technology to analyze soil microbial diversity under different gradients of petroleum pollution concentration and sequenced 16S amplicons by the soil, and the IlluminaMiSeq platform. The results showed that the microbial species in contaminated soil were more abundant than in clean soil, the main dominant bacterial groups were changed, and significantly more bacteria could degrade petroleum pollutants. At the same time, while the soil microbial diversity and community structure differed, it was not significant between different contamination levels and differed significantly from that of clean soil, consistent with the results of previous studies (Wu et al. 2020; Xu et al. 2021; Sun et al. 2021; Zhao et al. 2020).
At the genus level, Pantoea, Sphingomonas, Thiothrix, and Nocardioides were dominant in uncontaminated soil in high abundance. The intake of petroleum hydrocarbons resulted in a decrease in the abundance of Thiothrix, Sphingomonas, Gemmatimonas, Pseudomonas, Acinetobacter, Pedobacter, and the abundance of Pseudomonas, Acinetobacter, Pedobacter, and Massilia increased significantly. This may be because petroleum contaminants caused changes in water content, total organic carbon, total petroleum hydrocarbons, total carbon, total sulfur, and pH values, which are the limiting factors for the growth of the dominant genera (Liang et al. 2016), thus changing the relative abundance of Sphingomonas, Thiothrix, and Pseudomonas. The oleophilic bacteria genera in the contaminated soil in the typical oil extraction area in the Loess hilly area mainly include Pseudomonas, Acinetobacter, Pedobacter, Acinetobacter, Nocardioides, and Alkanindiges among others (Xu et al. 2021; Zhen et al. 2015).
These data can help us to better understand the evolutionary characteristics of various indigenous microorganisms in oil-contaminated soils in the region and provide theoretical reference and data support for remediation and treatment technologies of oil-contaminated soils in the region. In the future, several kinds of oleophilic indigenous bacteria need to be screened and domesticated from the contaminated soil in the study area to make bacteriological agents and conduct indoor simulation experiments to investigate and explore the applicability of indigenous oleophilic microorganisms in the bioremediation of petroleum-contaminated soil.
Availability of data and materials
The data were obtained by the authors.
References
Ajona M, Vasanthi P (2021) Bio-remediation of crude oil contaminated soil using recombinant native microbial strain. Environ Technol Innov 23(11):101635
Ali F, Nadeem MA, Habyarimana E et al (2020) Molecular characterization of genetic diversity and similarity centers of safflower accessions with ISSR markers. Braz J Bot 43(1):1–13
Chen FF, Cong X, Xiang JZ et al (2020) Application of Illumina MiSeq high-throughput sequencing technology to analyze the inter-rooted soil microbial diversity of Corydalis sativus. Hubei Agric Sci 59(17):6
Dong ZJ (2020) Study of soil contamination law by seepage of an oil pipeline in loess area. Eng Surv 48(5):6
Fan GP, Zhu HY, Hao XZ et al (2015) Electrokinetic remediation of an electroplating contaminated soil with different enhancing electrolytes. China Environ Sci 35(5):1458–1465
He YJ, Zhou KP, Rao YX et al (2021) Environmental risks of antibiotics in soil and bioremediation technology of contaminated soil. J Biol Eng 37(10):18
Iyobosa E, Zhu SF, Ning HJ et al (2021) Development of a robust bacterial consortium for petroleum hydrocarbon degradation. Fresenius Environ Bull 30(3):2356–2367
Kong LL, Wang XY, Gu MG et al (2021) Analysis of factors influencing enhanced biodegradation of petroleum hydrocarbons by biochar. Environ Sci Technol 44(3):8
Li J, Cao XT, Sui H et al (2017) Current status and prospect of research on remediation technology of petroleum-contaminated soil. J Pet (Pet Process) 033(005):811–833
Li JL, Liu GG, Cao SX et al (2011) Optimization of RAPD-PCR reaction system for partridge tea using one-way and orthogonal design. J Agron 1(4):8
Li Q, He LS, Wang YF et al (2021) Soil contamination characteristics and distribution of smelting industry sites in China. J Ecol Environ 30(3):10
Li SL, Liu GB, Xu MX (2009) Characteristics of petroleum pollution in soils and surface waters of oil fields in northern Shaanxi. Soil Water Conserv Res 16(5):4
Liang JF, Yang JK, Yang Y et al (2016) Relationship between bacterial community structure and environmental factors in oil-contaminated soils of the Karamay oilfield. J Microbiol 56(8):13
Liu JM, Peng JP (2001) One-way analysis of variance for biological experimental data. J Zool 36(6):4
Liu L, Wang DS, Zeng QS et al (2012) Homogeneous design experiments for microwave remediation of crude oil-contaminated soil. J Environ Eng 6(6):5
Liu MY, Ma JH, Li Y et al (2020) Effects of 16S rRNA gene high variation regions V4 and V3-V4 and sequencing depth on the analysis of bacterial flora in oil reservoirs. Microbiol Bull 2:10
Liu YP, Mao YF, Hu YL et al (2021) Effects of grass cultivation on soil microbial diversity, enzyme activity and carbon fraction in apple orchards. J Plant Nutr Fertil 27(10):14
Lusk R, Stene E, Banaei-Kashani F et al (2021) Aptardi predicts polyadenylation sites in sample-specific transcriptomes using high-throughput RNA sequencing and DNA sequence. Nat Commun 12(1):1652
Ma R, Jiang M, Li H et al (2020) Analysis of microbial diversity in northeastern bean paste based on high-throughput sequencing technology. Food Ind Sci Technol 41(12):6
Mambwe M, Kalebaila KK, Johnson T (2021) Remediation technologies for oil contaminated soil. Glob J Environ Sci Manag 1:1–20
Margesin R, Mmerle M, Tscherko D (2007) Microbial activity and community composition during bioremediation of diesel-oil-contaminated soil: effects of hydrocarbon concentration, fertilizers, and incubation time. Microb Ecol 53(2):259–269
Mu CR (1994) The impact of oil development on the ecological environment of the Loess Plateau. Geogr Stud 13(4):9
Pan F, Chen LH, Fa SJ et al (2012) A study on the transport performance of the petroleum contaminants in soil of the Longdong loess plateau. J Environ Sci 32(2):410–418
Sun J, Wang N, Chen HK et al (2021) Microbial community distribution characteristics of petroleum-contaminated soil. J Pet (Pet Process) 37(2):12
Sun YR, Tan L, Chen Y (2018) Soil contamination in typical oilfield areas in arid regions. Investigation and analysis of soil petroleum pollution in typical oilfield areas in arid regions. Environ Dev 30(11):2
Ueno A, Ito Y, Yamamoto Y et al (2010) Bacterial community changes in diesel oil contaminated soil microcosms biostimulated with Luria-Bertani medium or bioaugmented with a petroleum-degrading bacterium, Pseudomonas aeruginosa strain WatG. J Basic Microbiol 46(4):310–317
Wagh GA, Tiwari PD (2020) On the diversity and abundance of avian species from grassland and wetland areas of an industrial zone of tropical Maharashtra. Biosci Biotechnol Res Commun 13(2):770–780
Wang D, Ma B, Gao H et al (2020) Study on the remediation characteristics of petroleum-contaminated soil by microbial enhancement. J Agric Environ Sci 39(12):8
Wang J, Qi YC, Feng Q et al (2016) Relationship between the restoration of oil-contaminated soil vegetation and soil CAT activity on the Loess Plateau. Shaanxi Agric Sci 62(6):6
Wang JC, Jing MB, Duan CY et al (2017) Effects of petroleum hydrocarbon pollution on the biological and non-biological properties of soils on the Loess Plateau of Longdong. Soil Water Conserv Bull 37(1):8
Wang JD, Qu CT, Song SF (2021) Temperature-induced changes in the proteome of Pseudomonas aeruginosa during petroleum hydrocarbon degradation. Arch Microbiol 1:1–11
Wang R, Zhu J, Jin T et al (2019) Characterization of soil ammonia-oxidizing microbial abundance and community structure in rice fields under rice and shrimp crop model. J Plant Nutr Fertil 25(11):13
Wen GY, Zhao QZ, Ma HG et al (2017) Research and application of high-efficiency and low-cost bioremediation technology for petroleum-contaminated soil. China Saf Prod Sci Technol S1:5
Wu H, Song A, Zheng J et al (2020) Progress of functional genes for microbial degradation of petroleum hydrocarbons. Microbiol Bull 47(10):14
Xu YR, Wu ML, Wang L et al (2021) Changes in microbial populations and influencing factors of petroleum-contaminated soils in northern Shaanxi. China Environ Sci 41(9):11
Yang MQ, Li LM, Li C et al (2013) Microbial community structure and distribution characteristics of petroleum-contaminated soil. Environ Sci 34(2):6
Zhang GQ, Zhao P, Dong YX et al (2017) High-throughput sequencing analysis of the effect of environmental fertilizer enhancers on the change of soil fungal diversity in potato inter-rhizosphere. Microbiol Bull 44(11):8
Zhang J, Xu M (2022) Characteristics of soil bacterial community diversity in different vegetation types in loess hilly areas. J Ecol Rural Environ 38(2):225–235
Zhang L, Zhao Q, Wu WN et al (2018) Current status and prospect of bioremediation technology for petroleum-contaminated soil. Mod Chem Ind 38(1):5
Zhao MY, Wang X, Li FM, Guo SH et al (2020) Soil oil pollution and its microbial community characteristics in Liaohe oilfield. J Appl Ecol 31(12):10
Zhen L, Gu J, Hu T et al (2015) Microbial community structure and metabolic characteristics of petroleum-contaminated soils on the Loess Plateau. J Ecol 35(17):8
Zheng YM, He XS, Shan GC et al (2021) Effects of petroleum contamination on bacterial communities in field and its feedback mechanisms. Environ Sci Res 34(4):987-995
Zhu SS, Xia B, Hao WL et al (2020) Functional diversity of soil microbial communities on eroded slopes in loess areas. China Environ Sci 40(9):7
Acknowledgements
This work was supported by the projects of “Technology Innovation Leading Program of Shaanxi (Program No. 2021CGBX-03)” and Shaanxi Provincial Land Engineering Construction Group (Program No. DJNY2021-33).
Ethical statements
I certify that this manuscript is original, has not been published anywhere, and will not be submitted elsewhere for publication while being considered by the Annals of Microbiology. The study is not split up into several parts to increase the number of submissions and submitted to various journals or to one journal over time. No data have been fabricated or manipulated (including images) to support the conclusions. No data, text, or theories by others are presented as if they were our own.
The submission has been received explicitly from all co-authors, and authors whose names appear on the submission have contributed sufficiently to the scientific work and therefore share collective responsibility and accountability for the results.
Funding
This work was supported by the projects of “Technology Innovation Leading Program of Shaanxi (Program No. 2021CGBX-03)”, and Shaanxi Provincial Land Engineering Construction Group (Program No. DJNY2021-33, DJNY2022-37).
Author information
Authors and Affiliations
Contributions
LS guided the completion of this study, completed the first draft paper, and made revisions. WF guided the experimental methods and thesis writing. ZL performed experiments, recorded data, and data analysis. LY performed data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study did not violate ethics, and all participants agreed to publish the paper.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, L., Liu, Z., Yang, L. et al. Effects of oil pollution on soil microbial diversity in the Loess hilly areas, China. Ann Microbiol 72, 26 (2022). https://doi.org/10.1186/s13213-022-01683-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13213-022-01683-7