Abstract
Background
Methylation of serotonin-related genes has been proposed as a plausible gene-by-environment link which may mediate environmental stress, depressive and anxiety symptoms. DNA methylation is often measured in blood cells, but little is known about the association between this peripheral epigenetic modification and brain serotonergic architecture. Here, we evaluated the association between whole-blood-derived methylation of four CpG sites in the serotonin transporter (SLC6A4) and six CpG sites of the tryptophan hydroxylase 2 (TPH2) gene and in-vivo brain levels of serotonin transporter (5-HTT) and serotonin 4 receptor (5-HT4) in a cohort of healthy individuals (N = 254) and, for 5-HT4, in a cohort of unmedicated patients with depression (N = 90). To do so, we quantified SLC6A4/TPH2 methylation using bisulfite pyrosequencing and estimated brain 5-HT4 and 5-HTT levels using positron emission tomography. In addition, we explored the association between SLC6A4 and TPH2 methylation and measures of early life and recent stress, depressive and anxiety symptoms on 297 healthy individuals.
Results
We found no statistically significant association between peripheral DNA methylation and brain markers of serotonergic neurotransmission in patients with depression or in healthy individuals. In addition, although SLC6A4 CpG2 (chr17:30,236,083) methylation was marginally associated with the parental bonding inventory overprotection score in the healthy cohort, statistical significance did not remain after accounting for blood cell heterogeneity.
Conclusions
We suggest that findings on peripheral DNA methylation in the context of brain serotonin-related features should be interpreted with caution. More studies are needed to rule out a role of SLC6A4 and TPH2 methylation as biomarkers for environmental stress, depressive or anxiety symptoms.
Similar content being viewed by others
Background
Most psychiatric disorders, including major depressive disorder (MDD), arise from a complex etiology, with contributions from genetic and environmental factors. The serotonin system mediates a variety of different functions from the very early stages of development and throughout life, including cognition, mood and sleep as well as adaptation to environmental challenges [1,2,3,4]. For instance, serotonin-mediated neuroplasticity has been suggested to allow us to adapt to the ever-changing environment [5]. In this case, alterations in serotonin function might translate into resilience or vulnerability to MDD [2, 6].
DNA methylation of genes coding for key regulators of the serotonin system, such as the serotonin transporter (SLC6A4) and the tryptophan hydroxylase 2 gene (TPH2), has been proposed as a possible gene-by-environment mechanism involved in several psychiatric disorders, including MDD [7, 8]. However, DNA methylation is often measured in peripheral samples (e.g. blood or saliva) and little is known of the effect of this modification on in-vivo brain serotonin transmission. In this study, we investigated the association between methylation of peripheral SLC6A4 and TPH2 and brain proxies of serotonin transmission measured with in-vivo positron emission tomography (PET) imaging.
The serotonin transporter (5-HTT) and tryptophan hydroxylase 2 (TPH2) critically shape serotonin signalling by regulating serotonin levels. Specifically, 5-HTT regulates synaptic levels of serotonin available for neurotransmission and is the main target of selective serotonin reuptake inhibitors (SSRI), the most widely used class of antidepressant medications. TPH2 is the rate-limiting enzyme for serotonin synthesis in the brain [9], thereby directly affecting presynaptic serotonin levels.
A role of SLC6A4 in gene-by-environment interaction was initially described by Caspi and colleagues [10], reporting that 5-HTTLPR s-carriers, who had lower SLC6A4 expression, were more vulnerable to stress in terms of developing depressive episodes when experiencing stressful life events. Nonetheless, these findings have not been replicated by all larger studies [11, 12], suggesting that 5-HTTLPR per se may not be as relevant to MDD or anxiety-related traits as previously thought. Instead, a combination of genetic and epigenetic factors may affect SLC6A4 gene expression levels [13] in a way that may be relevant to the development of psychopathology [14].
Over the last two decades, several studies have pointed to a possible role of DNA methylation levels in the transcriptional control region of SLC6A4 as a marker of gene-by-environment interaction [7, 15,16,17]. Specifically, alterations in SLC6A4 methylation have been associated with recent [15] and early life stress [7, 18], depressive symptoms [19], panic disorder [16] and likelihood to respond positively to antidepressant treatment [20, 21], although the relation with depressive symptoms and antidepressant treatment outcome was not confirmed by all studies [20, 22].
More recently, methylation of TPH2 gene has also been suggested as a biomarker for vulnerability to depression and antidepressant treatment outcome [23, 24].
DNA methylation at cytosine-guanine dinucleotides (CpG) is a common epigenetic modification which can affect gene expression in response to environmental cues [25]. Early studies reported an association between SLC6A4 methylation and 5-HTT mRNA levels measured in lymphocytes [13] and peripheral whole blood [26, 27]. However, it is not known whether such altered peripheral 5-HTT mRNA levels also correlate with brain 5-HTT protein levels. Given the fundamental role of 5-HTT and TPH2 in serotonin neurotransmission, understanding whether peripheral SLC6A4 or TPH2 methylation mirrors serotonin brain architecture is essential to interpret previous findings and to shed light on the role of peripheral methylation in the context of health and disease, e.g. psychiatric disorders.
Indeed, it is important to note that DNA methylation is cell-type specific [28]. However, as brain tissue of living human participants is mostly unavailable for biomarker assessment, blood and saliva are the most used tissue types for the investigation of DNA methylation. Peripheral blood and postmortem brain DNA methylation partially correlate at multiple CpG sites, but there is not a perfect correspondence between the two tissues [29,30,31]. Evaluating DNA methylation associations with in vivo brain serotonin markers allows to estimate its relevance as a peripheral marker of serotonin neurotransmission. However, to our knowledge, no study has investigated whether peripheral methylation of SLC6A4 or TPH2 is associated with brain levels of 5-HTT or with other markers of serotonin neurotransmission, such as serotonin 4 receptor (5-HT4), a post-synaptic serotonin receptor that has been proposed as a biomarker for brain serotonin tonus [32]. Only one study reported an association between SLC6A4 promoter methylation and brain serotonin synthesis measured in terms of brain tryptophan levels [27].
PET imaging allows quantification of serotonin system protein levels in the living brain [33, 34]. In this study, we used PET scans of 254 healthy participants and 90 patients with MDD to determine the relation between peripheral SLC6A4 and TPH2 methylation and two key features of the serotonergic brain signalling system, i.e. 5-HTT and 5-HT4, imaged with combined with [11C]DASB and [11C]SB207145 PET radiotracers, respectively. Both the 5-HTT and 5-HT4 are known to play a role in healthy brain function and in MDD pathology and can be considered as key markers for serotonin neurotransmission [35,36,37,38].
Furthermore, as primary sensitivity analyses, we evaluated the association between DNA methylation and self-reported early life stress and stressful life events, as well as state measures of perceived stress and anxiety and depressive symptoms in 297 healthy participants.
Finally, blood is a heterogeneous tissue containing different cell types. Interindividual differences in blood cell proportions can be a source of bias on DNA methylation measurements carried out on whole blood [39], hindering comparability between individuals. Nonetheless, while epigenome-wide studies routinely correct for blood cell proportions [40], most of previous studies linking SLC6A4 and TPH2 methylation to environmental stress [15, 41] or psychiatric conditions [7, 16, 21, 23] did not account for blood cells proportions. Thus, we used blood cell counts to estimate blood cell proportions in a subgroup of participants for whom this information was available. Then, we included cell proportions in all our statistical models as secondary sensitivity analyses.
Methods
Participants
All participants included in this study were recruited as part of neuroimaging projects conducted at Neurobiology Research Unit, Copenhagen University Hospital Rigshospitalet, in compliance with the Declaration of Helsinki and Good Clinical Practice guidelines. An overview of the methods is depicted in Fig. 1.
Overview of the data and methods used in this study. a, b, c, d depict the primary analyses, in which a latent variable model was used to determine the association between peripheral TPH2 and SLC6A4 methylation and brain levels of 5-HTT and 5-HT4. e, f, g describe the sensitivity analyses evaluating the association between SLC6A4/TPH2 methylation and measures of environmental stress, depressive and anxiety symptoms. h and i show sensitivity analyses used to evaluate potential influence of blood cell proportions in the A-G analyses. Abbreviations: 5-HT4: serotonin 4 receptor; 5-HTT: serotonin transporter; CpG: CpG site; TSS: transcription start site; SLE: stressful life events; PBI: parental bonding inventory; BDI: Beck’s depressive index; GAD10: generalized anxiety disorder 10-item; CATS: childhood abuse trauma scale; HAMD6: Hamilton depressive rating scale 6; PSS: perceived stress scale; CpGLV: latent variable including all CpG methylation values
Healthy Cohort
We included data of healthy volunteers from the Cimbi database and biobank [42]. The data was included based on the following criteria: (1) availability of [11C]DASB PET or [11C]SB207145 PET; (2) availability of whole blood or buffy coat samples matching scan date (blood samples drawn maximum one week before or after the PET scan were also included for N = 3) and (3) self-identification with European ancestry. Before inclusion in any of the original studies, all participants were screened for psychiatric disorders and underwent a physical and neurological examination. Participants with a history of psychiatric illness or current use of psychotropic drugs or drugs potentially affecting PET measurements were excluded.
We identified a cohort of 142 participants with [11C]DASB PET scans and a cohort of 112 participants with [11C]SB207145 PET scans. Demographic data relative to the cohorts included in the study are depicted in Table 1.
To evaluate the potential association between DNA methylation and early life stress history or state measures of perceived stress, depressive or anxiety symptoms, data from an additional 43 healthy participants without PET scans was available, resulting in a total of 297 healthy participants. Demographics for the participants included in all analyses are depicted in Table S1.
MDD patient cohort
We included baseline data from 90 unmedicated patients with moderate to severe unipolar MDD that were originally part of the NeuroPharm-1 study [43], an open-label, non-randomized longitudinal clinical trial. Patients were included based on the availability of both blood samples and [11C]SB207145 PET scans that were collected no more than one week apart.
The primary outcome of the trial involved measures of molecular neuroimaging and cognitive functions and is described in previous publications [36, 43, 44]. Shortly, previously unmedicated patients with MDD were recruited for an open-label clinical trial aiming to uncover biomarkers predicting clinical outcome after 12 weeks of antidepressant treatment. In this study, we included blood samples, PET scans and psychometric data collected at baseline. Analyses carried out in this study involving SLC6A4 are planned secondary analyses. Analysis including TPH2 are unplanned exploratory analyses. Participants were evaluated in face-to-face interviews and diagnosed by a certified psychiatrist. Individuals between 18–65 years of age, scoring > 17 in the Hamilton Depression Rating Scale 17 items (HAMD17)[45] and who were unmedicated for at least two months before the start of the trial were included in the study. Additional details about the trial as well as inclusion and exclusion criteria are specified in [43].
DNA methylation analysis
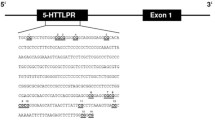
SLC6A4 methylation percentages were estimated at four CpG sites (Table S2) that were previously linked to clinical phenotypes, including depressive symptoms [7, 16, 26, 46], early-life adversities [7], recent environmental stress [15], antidepressant treatment outcome [20, 21] and panic disorder [16]. TPH2 methylation was estimated at 6 CpG sites (Table S2) based on previous studies showing an association with gene expression (13, 26, 28), early life stress, depressive symptoms, antidepressant treatment outcome [23, 24, 47] and attention deficit hyperactivity disorder [48].
Genomic DNA was isolated from peripheral blood cells from whole blood or buffy coat samples that were stored at − 20 °C (MDD patient cohort) or − 80 °C (healthy cohort) in EDTA tubes. DNA was purified using the FlexiGene Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. 500 ng DNA of each sample were bisulfite-converted using the EpiTect 96 Bisulfite Kit (Qiagen). The sequence of interest was amplified via polymerase chain reaction (PCR) using the PyroMark PCR Kit (Qiagen) and a forward (F) and a reverse (R) biotinylated primer (Table S3). The quality of PCR amplification was visually evaluated using gel electrophoresis. The target DNA sequence was isolated and then sequenced using the PyroMark Q96 ID (Qiagen) pyrosequencing system, with target-specific primers (Table S3). CpG methylation rates (in %), pyrograms and quality reports were obtained using the PyroMark software (Qiagen). Analyses were run in duplicates and pairs of duplicates differing more than 3% from each other were excluded from the analyses. Average DNA methylation value between each pair of duplicates was used for statistical analyses. Pyrograms and quality reports provided by PyroMark were used to quality check the data. Commercially available (Epitect PCR Control DNA Set, Qiagen) fully methylated, fully non-methylated and 50%-methylated DNA samples as well as DNase free H2O were included in all experiments as controls. Methylation data of two samples for SLC6A4 and three samples for TPH2 were excluded due to failed bisulfite conversion, as indicated by the PyroMark software.
Genotyping
All samples were genotyped for SLC6A4 5-HTTLPR and rs25531, TPH2 rs4570625, BDNF rs6265 and MAOA rs1137070 polymorphisms. Genotyping for SLC6A4 5-HTTLPR and rs25531, BDNF rs6265 and MAOA rs1137070 was performed as previously described [35, 49,50,51]. Hardy–Weinberg equilibrium was tested using Chi-squared test in R. Table 1 and S1 show allele frequencies within all cohorts.
PET and MR data acquisition and processing
The acquisition, preprocessing and quantification of [11C]SB207145 and [11C]DASB PET and MR images has been previously reported [37, 50, 52]. For each participant, both PET and concomitant MR scans were acquired. MR scans coregistered to PET were used to delineate brain regions and quantify regional PET signal.
Shortly, all participants were scanned for a 120-min ([11C]SB207145) or a 90-min ([11C]DASB) dynamic scan after bolus injection of the respective radioligand. Two different PET scanners were used for data collection: a High-resolution Research Tomography (HRRT) PET scanner (CTI/Siemens) with an approximate in-plane resolution of 2 mm, or an 18-ring GE-Advance PET scanner (General Electric, Milwaukee, USA) with an approximate in-plane resolution of 6 mm. T1-weighted MPRage images were acquired using three different Siemens 3-Tesla magnetic resonance (MR) scanners: Prisma, Trio or Verio. Regions of interest (ROI) were automatically delineated using PVElab and the individual T1-weigthed images [53]. Mean time-activity curves for average grey matter voxels in each hemisphere was determined using the Simplified Reference Tissue Model for [11C]SB207145 scans and multilinear reference tissue model (MRTM/MRTM2) for [11C]DASB scans. Cerebellum (except for vermis) was used as a reference region for all scans. The non-displaceable binding potential (binding or BPND) was used as an outcome measure of tracer binding (and therefore as an estimate of 5-HTT and 5-HT4 levels) for both tracers.
Statistical analyses
All statistical analyses were conducted in R v4.1.2 [54].
Primary analyses
The association between SLC6A4/TPH2 methylation and 5-HTT or 5-HT4 was evaluated using three different linear latent variable models (LVM): one for healthy controls with DASB scans (5-HTT binding), one for healthy controls with SB scans (5-HT4 binding) and one for MDD patients with SB scans. LVM is a type of multivariate linear regression that allows to model associations between a variable of interest and the shared variance of a set of inter-correlated variables (e.g. 5-HTT or 5-HT4 binding in different brain regions).
Regions of interest (ROI) for the 5-HTT LVM were chosen based on 5-HTT distribution in the human brain [55] and comprised caudate, amygdala, hippocampus, putamen, thalamus, midbrain and neocortex. Similarly, ROIs for the 5-HT4 LVMs include caudate, putamen, hippocampus and neocortex, reflecting brain regions across low, moderate to high density of 5-HT4 receptor in these areas [55] and aligning with previous findings investigating the 5HT4 receptor system and MDD [32, 56].
Analyses were carried out in R and the LVMs were modelled using the lava v 1.6.10 [57] package. First, the shared correlations of regional 5-HT4 or 5-HTT binding were modelled into a latent variable for each model (referred to as 5-HT4LV or 5-HTTLV respectively). Next, SLC6A4 CpG1-CpG4 or TPH2 CpG1-6 methylation and the covariate effects were modelled on the 5-HTTLV. In all models, covariates included age, sex, PET scanner type (Advance vs HRRT) and MR scanner type (Prisma vs Trio vs Verio). 5-HTTLPR/rs25531 genotype was included in the statistical models including SLC6A4 methylation based on previous studies suggesting a combined effect of genotype and DNA methylation on 5-HTT transcription [13, 17]. Similarly, TPH2 rs4570625 was included in the models evaluating TPH2 methylation [48]. Models evaluating associations between DNA methylation and 5-HT4 binding also included information of 5-HTTLPR/rs25531 and BDNF rs6265, based on previous findings [58]. In addition, BDNF rs6265 and MAOA rs1137070 genotype information, which have been previously shown to affect 5-HTT [49, 50, 58], were included in a separate model as sensitivity analyses, as information for the latter genotype was not available for all subjects. Region-specific effects of each CpG site were evaluated as the product of the CpG effect on the latent variable multiplied by the loading of each region on the latent variable and were used as a measure of effect sizes.
PET and MR scanner type were modelled as region-specific effects, based on previous findings [58]. Additional covariance links were identified using an iterative procedure where score tests are used to detect model misspecification. P-values for these score test were adjusted using Benjamini–Hochberg [57].
The LVMs used for primary analyses are graphically represented in Figs. 2 and 3.
Associations between peripheral SLC6A4 methylation and brain 5-HTT binding (a) or 5-HT4 binding (b). Blue dashed boxes depict the SLC6A4 CpG sites and the covariates included in the model. The light blue boxes indicate the CpG and covariate effects on the latent variable (5-HTTLV or 5-HT4LV). Dark grey boxes to the right represent the observed 5-HTT or 5-HT4 binding in the brain regions of interest. β values refer to the parameter estimates; they are reported either with their respective p-values or with their 95% confidence intervals. Dashed arrows connecting brain regions show interregional correlations, while dashed circles on the brain regions show error estimates. For representation purposes, PET and MR scanner covariates are not reported in the a and b models. Similarly, although included in the 5-HT4 latent variable model, 5-HTTLPR/rs25531 and BDNF rs6265 genotypes are not reported in (b). Scatter plots in c and d depict the relation between SLC6A4 methylation and 5-HTTLV or 5-HT4LV in healthy controls (c, d), while the relation between SLC6A4 methylation and 5-HT4 binding in patients with MDD is shown in (e)
Associations between peripheral TPH2 DNA methylation and brain 5-HTT binding a or 5-HT4 binding b in the healthy cohort. Orange dashed boxes to the left depict the TPH2 CpG sites and the covariates included in the model. Rs45706210 stands for TPH2 rs45706210 G/T SNP. For representation purposes, PET and MR scanner covariates are not reported in the a and b models. Similarly, although included in the 5-HT4 latent variable model, 5-HTTLPR/rs25531 and BDNF rs6265 genotypes are not reported in (b). Scatter plots in c and d depict the relation between TPH2 methylation and 5-HTTLV or 5-HT4LV in healthy controls (c, d), while the relation between TPH2 methylation and 5-HT4 binding in patients with MDD is showed in (e)
Caudate was used as a reference region in all LVMs. Thus, covariate effects can be interpreted as effects on caudate binding. Statistical significance was set at p < 0.05 for all the statistical models.
Sensitivity analyses I: DNA methylation vs measures of environmental stress, depressive and anxiety symptoms
Multiple linear regression models were used to explore the relation between methylation of each CpG site and measures of environmental stress in both healthy participants and MDD patients. Associations with depressive and anxiety symptoms were also evaluated in the MDD patients. Data of all the healthy controls were pooled together with data of 58 additional healthy participants, for a total of N = 297 (Table S1). Data from the MDD patients were the same as those used for the LVM analyses. The stressful life events (SLE) questionnaire was used as an estimate of both lifetime (total SLE score) and recent stress (recent SLE score) in the healthy cohort. The parental bonding inventory (PBI) was used as a proxy estimate of early life stress in both the healthy and the MDD cohorts. Scores from both parents were combined into a measure for the “care” (PBI care score) and one for the “overprotection” (PBI overprotection score) subscales. In addition, models exploring the association between SLC6A4 or TPH2 DNA methylation and the following measurements were carried out in the MDD cohort: (1) Beck’s Depression Inventory (BDI) indexing recent depressive symptoms; (2) childhood abuse trauma scale (CATS) as a measure of early life stress; (3) generalized anxiety disorder 10-item (GAD10); (4) Hamilton depression rating scale 6 item (HAMD6) indexing current depressive symptoms; (5) perceived stress scale (PSS) indexing recent stress symptoms.
All statistical models included age, sex and genotype (5-HTTLPR in the case of SLC6A4 or rs4570625 for TPH2) as covariates. Bonferroni correction for four and six tests was applied for analyses including SLC6A4 and TPH2 data, respectively (SLC6A4: p = 0.01; TPH2: p = 0.008).
Sensitivity analyses II: analyses accounting for cell type proportions
Sensitivity analyses were conducted to evaluate whether different blood cell type proportions affected the associations evaluated in the primary analyses and in the primary sensitivity analyses. Blood cell counts information was available only for a subset of the total participants used in the analyses (Table 1; panel H of Fig. 1). Corrections were done for lymphocytes, monocytes, granulocyte precursors and neutrophils proportions. The term granulocyte precursors used here refers to the sum of granulocyte precursors metamyelocytes, myelocytes and promyelocytes. Cell proportions were calculated by dividing the cell counts of each cell type by the number of leukocytes, multiplied by 100.
For the models evaluating the association with brain 5-HTT or 5-HT4 levels, the correction for cell type involved first modelling two latent variables, one including the shared correlations among DNA methylation across the four CpG sites (CpGLV) and one including the shared correlations of 5-HTT (5-HTTLV) or 5-HT4 binding (5-HT4LV) across model-specific ROIs. CpGLV was adjusted for age, sex, genotype and cell proportions by regressing out all cell proportions but neutrophils proportions (CpGLV+cells). Finally, the covariance between CpGLV+cells and 5-HTTLV or 5-HT4LV was estimated (panel I of Fig. 1).
To account for cell type proportions when evaluating the association between peripheral SLC6A4 or TPH2 DNA methylation and measures of environmental stress, depressive or anxiety symptoms, LVMs were used instead of linear regressions. For a given gene, a latent variable reflecting DNA methylation at all CpG sites was modelled (CpGLV) and adjusted for cell types (CpGLV+cells). Next, for every psychometric measurement, LVMs containing CpGLV+cells were regressed out on the psychometric score.
In all models including CpGLV+cells, the effect of cell types was tested using a likelihood ratio test between the LVM including CpGLV (without adjustment for cell proportions) and the corresponding LVM including CpGLV+cells (adjusted for cell proportions). Whenever significant, cell type specific effects estimated by the LVM were reported without adjustment for multiple comparisons.
Results
Genotyping
Alleles were in Hardy–Weinberg equilibrium (p > 0.1) in all cohorts used for statistical analyses (5-HTT, MDD and healthy participants used in sensitivity analyses I) except for rs4570625 in the 5-HT4 cohort (χ2: 6.12; p = 0.01). However, rs4570625 did not deviate from Hardy–Weinberg equilibrium for the whole population used in this study (N = 389; χ2: 0.34; p = 0.56), suggesting that the lack of equilibrium might be due to chance and not to biases in genotyping.
Association between peripheral DNA methylation and brain serotonergic markers
Loadings, i.e. parameters evaluating the association between the latent variable and the 5-HTT or 5-HT4 binding values, were all significantly different from 0 (all p < 10–4), indicating evidence for shared variance among the 5-HTT and 5-HT4 in the respective ROIs.
The LVMs did not reveal a statistically significant association between SLC6A4 or TPH2 methylation and 5-HTTLV or 5-HT4LV in the healthy cohort nor the cohort of MDD patients (unadjusted p-values ranged between 0.06 and 0.97; Table 2). A graphical representation of the LVMs including the results is reported in Figs. 2 and 3.
In line with previous studies [49, 50, 58, 59] based on the same cohort, we observed: (1) a negative association between age and 5-HTTLV and 5-HT4LV in the healthy cohort (p < 0.01) but no association between age and 5-HT4 binding in the MDD cohort; (2) a non-significant effect of 5-HTTLPR on 5-HTTLV; (3) an association between MAOA rs1137070 and 5-HTTLV (MAOA T- carriers vs CC, β: 0.1, 95% CI: [0.02; 0.18], p = 0.01) and between neocortex binding and BDNF rs6265 (with lower subcortical binding for met-carriers, estimate (β): − 0.02, 95% CI: [− 0.04; 0.01], p = 0.005) in the subset of the sample with this information available (N = 130), 4) higher 5-HT4 binding in male compared to female participants. Contrary to previous observations based on a subgroup (N = 68/112) of the participants included in this study (Fisher et al. 2015: β: 0.070, 95% CI: [0.018; 0.122], p = 0.008), we did not observe a statistically significant association between 5-HTTLPR or BDNF rs6265 genotypes and 5-HT4LV. In addition, we did not find any association between TPH2 rs4570625 and 5-HTTLV or 5-HT4LV (Table 2). Estimated effect sizes and respective 95% CI for effects of each CpG site on each brain region are reported in Tables S4 and S5 for models including SLC6A4 and TPH2 respectively. Compared to age, which is known to affect 5-HTT and 5-HT4 binding by about 9% and 1% per decade, respectively [59, 60], the effect sizes of our study were minimal. The largest effect sizes in our dataset indicated that 5-HT4 binding decreases by 0.14% for each one-unit increase in SLC6A4 methylation (Table S4) and by 0.24% for each one-unit increase of TPH2 methylation (Table S5).
DNA methylation and measures of environmental stress
Results from multiple linear regressions on all cohorts are reported in Table S6 and S7. Among all statistical tests, only three associations reached the threshold for statistical significance before correction for multiple comparisons, and only the association between SLC6A4 CpG2 and PBI overprotection item remain statistically significant at the 5% level after Bonferroni correction (β: − 0.83; pUNC = 0.01; 95% CI: − 1.48; − 0.19).
Corrections for cell type
Loadings of DNA methylation at single CpGs onto CpGLV were all significantly different from 0 (all p < 0.01). Likewise, loadings of regional 5-HTT or 5-HT4 binding significantly loaded onto their corresponding latent variables 5-HTTLV or 5-HT4LV (all p < 10–11).
Likelihood ratio tests showed an improved model fit when including cell proportions in all the healthy cohorts (all p < 0.01). However, in the MDD cohort only the models evaluating the association between TPH2 methylation and BDI, HAMD6, PSS and GAD10 showed improved model fit after adding cell proportions.
Lymphocytes proportion was significantly associated with CpGLV in the TPH2 models including the healthy cohort (DASB: β: 0.017, p = 0.017, 95% CI: [0.004; 0.04]; SB: β: 0.016, p = 0.019, 95%CI: [0.003; 0.03]) and both in the SLC6A4 and TPH2 models based on the MDD patients cohort (SLC6A4: β: 0.014, p = 0.03, 95% CI: [0.002; 0.03]; TPH2: β: 0.01; p = 0.01; 95% CI: [0.002; 0.02]). No statistically significant association was found between CpGLV and any cell type, age or sex in the SLC6A4 model.
Accounting for blood cells proportion did not affect the conclusions about the associations between SLC6A4 or TPH2 methylation and 5-HTTLV or 5-HT4LV in the healthy cohort nor the cohort of MDD patients (Table 2), as all p-values were greater or equal to 0.08 (Table 3).
All models evaluating the association between SLC6A4/TPH2 methylation and measures of early stress, anxiety or depressive symptoms and including cells proportions showed a significant association between lymphocytes proportions and CpGLV (Table S8). Contrarily, granulocyte precursors were marginally statistically significantly associated only with SLC6A4 CpGLV in the model including healthy participants (Table S8). The other cell types considered showed no association. Age was statistically significantly associated with TPH2 CpGLV but not with SLC6A4 CpGLV. Before adjusting for multiple comparisons, TPH2 CpGLV was associated with sex in the healthy participants but not in the MDD participants, with higher TPH2 CpGLV values in males compared to females (Table S8).
Associations between CpGLV+cells and measures of environmental stress or mood or anxiety symptoms are depicted in Table S9 and showed no statistically significant association.
Discussion
In this study we found no statistically significant associations between peripheral DNA methylation of two key regulatory genes of serotonin neurotransmission (SLC6A4 and TPH2) and brain levels of 5-HTT and 5-HT4 in a cohort of healthy participants or 5-HT4 in a cohort of unmedicated patients with MDD.
Previous evidence supports an association between the CpG sites observed in our study and psychopathological features [7, 16, 48]. However, little is known about how peripheral DNA methylation of serotonin genes maps onto the brain serotonergic architecture. Only one study reported that increased SLC6A4 methylation was associated with reduced in-vivo TPH2 brain levels in a cohort of adult males that experienced childhood aggression [27]. Nonetheless, the study was based on a relatively small cohort (N = 25) that experienced high childhood aggression while our study, although based on a notably larger cohort, includes participants that did not experience extreme childhood traumas.
The lack of an association observed in our study should be considered also in light of the intricate nature of gene regulation. First, DNA methylation levels can differ across tissues. Previous epigenome-wide association studies reported that methylation of some CpG sites correlate between peripheral blood cells and entorhinal cortex, cerebellum, superior temporal gyrus and prefrontal cortex in postmortem brains of elderly adults [29, 61]. To evaluate a correspondence of DNA methylation between the two tissues, we consulted the online database created by Hannon et al. (2015) and found that only SLC6A4 CpG4 methylation levels correlate with methylation in entorhinal cortex (pUNC = 0.02) and superior temporal gyrus (pUNC = 0.04). SLC6A4 CpG1 and TPH2 CpG2 did not show any correlation, while information on the other CpG sites included in our study or other brain regions was not available in their database, so we cannot exclude a correspondence between the two tissues at other sites.
Second, even in the case DNA methylation was consistent across tissues, different transcription factors might interact differently with similar DNA methylation patterns in different tissues [62]. Thus, assuming similar DNA methylation levels between the two tissues, it is not known whether SLC6A4 or TPH2 expression would be affected in the brain in the same way that it is known to be affected in peripheral blood [13, 26, 27, 63]. Third, gene expression does not always directly correspond to protein levels as post-transcriptional and post-translational modifications can affect protein levels and function, and this notion seems to be true for both genes [55, 64,65,66,67]. This might also help explain why we did not observe any association between 5-HTTLPR or TPH2 rs4570625, which are polymorphisms known to affect SLC6A4 and TPH2 expression, and 5-HTT or 5-HT4 levels, which is in line with former studies [50, 68].
Nonetheless, most of this evidence is based on studies in adult individuals and we cannot rule out an effect of genetic variation or DNA methylation within serotonergic genes on early brain development, which is critically driven by serotonin transmission [69, 70]. Indeed, while brain 5-HTT and 5-HT4 levels may vary substantially throughout the lifespan [35] and in response to environmental changes [71,72,73,74], DNA methylation remains stable at about half of the total CpG sites after the first years of life [75, 76]. Using the online database provided by Mulder et al. [76], we observed no change in DNA methylation at SLC6A4 CpG1 or CpG4 or TPH2 CpG2 over the first 17 years of life of healthy individuals, although information on the other CpG sites relevant to our study was not provided. Longitudinal study designs with methylation sampling and PET imaging would allow to better understand if this was the case.
Notably, the lack of association between peripheral DNA methylation and adult brain levels of serotonergic markers does not necessarily imply that SLC6A4 or TPH2 methylation cannot be used as an informative biomarker for mental health. Instead, it might reflect peripheral alterations, e.g. of the immune system which can be critical for mental health. Altered immune function has been described in individuals who have experienced early life stress [77] and stress-related disorders, including depression [78]. Likewise, previous findings relating SLC6A4 and TPH2 methylation to measures of early life stress [7, 79] or depressive symptoms [7] might reflect alterations in peripheral immune function rather than in the brain serotonergic transmission.
In this regard, our sensitivity analyses revealed a borderline significant association between SLC6A4 CpG2 methylation at these genes and the parental bonding inventory (PBI) overprotection subscale, i.e. a proxy for suboptimal early social environment. However, this association was no longer statistically significant after including cell proportions in the model. Nonetheless, it is relevant to note that information of blood cells counts was available only for a subset of the total participants used for the sensitivity analyses, so such changes may be due to lower statistical power instead of the removal of unwanted variance. Thus, we suggest that this finding should be interpreted only if replicated in other cohorts.
Our study is the largest (N = 297) to date investigating the association between SLC6A4/TPH2 DNA methylation and early life stress in a healthy cohort. In line with our study, the second largest study based on a cohort of healthy participants (N = 133) [41] reported no association between SLC6A4 methylation and measures of early life stress. Our measurements for early life stress were PBI and SLE, which are based on retrospective self-reports and may not be as sensitive as other measurements in capturing early life stress. However, Wankerl et al. [41] did not find an association with early life stress, although both information of early-life stress reported by the participants’ mothers and self-reported were used. Thus, we can speculate that alterations in DNA methylation levels might only become detectable in case of more extreme (early) environmental stressors or in pathological conditions. Indeed, most previous studies linking peripheral SLC6A4 or TPH2 methylation to stress-related phenotypes were based on patients with mood disorders [7, 16] or individuals who were exposed to intense environmental stress [15, 27]. Our MDD cohort is smaller (N = 90) than some of those previously investigated (N > 100) [7, 16], so the results of our sensitivity analyses might be ascribable to a lack of power. Alternatively, the psychometric measurements used in our study might not be as sensitive at capturing early life stress as those used in other studies.
Importantly, the relation between SLC6A4 and TPH2 methylation and early life stress or brain levels of 5-HT4 or 5-HTT might also be affected by other environmental factors that were not considered in our study. For example, smoking [80], alcohol consumption [77, 81] or exposure to air pollutants [82] are known to affect gene expression through epigenetic modifications such as DNA methylation. Thus, we cannot exclude that future study designs including extreme exposure groups may inform on the potential effects of such environmental factors on a link between DNA methylation and serotonergic brain architecture.
In line with previous studies, we found that 5-HT4 binding was higher in men compared to women in healthy participants cohort [59] but not in the MDD cohort [83]. Previous studies investigating SLC6A4 and TPH2 methylation suggests that DNA methylation levels might be affected by sex [79, 84]. However, in our dataset we did not observe any conclusive effects of sex on neither gene, except for a trend in TPH2 CpGLV in the healthy participants. Notably, we observed it only before correcting for multiple comparisons, which we therefore interpret with caution. We did not observe the same effect on the MDD cohort.
It is important to mention some strengths of the present study compared with previous literature. First, it is based on the currently largest dataset in the world for brain molecular imaging for 5-HTT and 5-HT4. This also allows us to validate previous findings based on the same cohort such as in the case of BDNF rs6265 and 5-HT4 [58], which was initially found in a subset (N = 68) of the participants included in this cohort but could not be replicated in the more recent and larger current cohort (N = 112). Second, it includes both healthy participants and MDD patients, allowing us to investigate potential associations unique to healthy or pathological states; third, in our analyses we included blood cells proportions, which has rarely been done in former studies evaluating methylation at SLC6A4 or TPH2 and can be the main driver of methylation variability across individuals [39, 85].
However, this study also comes with several limitations. First, we examined four and six CpG sites for SLC6A4 and TPH2 respectively, which is only a small fraction of the total CpG sites in these genes and even smaller of those across the genome. An epigenome-wide exploration would be more informative, although a much larger sample size would be needed to capture potential peripheral epigenetic signatures associated with brain serotonergic transmission. For this reason, we chose a candidate epigenetic marker strategy for this study. Second, we considered only 5-HTT and 5-HT4 as proxies for serotonergic neurotransmission. Although this was based on previous evidence, there are many more contributors to serotonergic neurotransmission. In addition, in the MDD cohort we were only able to explore the association between DNA methylation and 5-HT4 levels since we did not have data on 5-HTT brain binding from this group. Alterations in brain 5-HTT levels have been reported in MDD patients [86] and future studies are needed to explore the relation between peripheral SLC6A4 methylation and brain 5-HTT levels in MDD. Nonetheless, preclinical and clinical studies show that 5-HT4 levels vary in response to serotonin levels [32, 87, 88] and in-vivo 5-HT4 levels deviate between healthy participants and MDD patients [36], pointing to the relevance of this target as a proxy for serotonin transmission in the context of MDD.
Third, although we included cell proportions in our sensitivity analyses, we could not take into account all blood cells subtypes (e.g. lymphocyte subtypes such as CD4+ or CD8+) but only the broader classes or cell types (monocytes, lymphocytes, neutrophils and granulocytes precursors) that are commonly evaluated in clinical routine. Thus, although we have accounted for some of the variance deriving from blood cell composition, we cannot assume that our analyses have accounted for all the variance.
Fourth, we could not account for the timing of possible traumas experienced by our participants. Cumulative evidence shows that the timing at which environmental stress was experienced can differentially affect DNA methylation [89, 90] as well as the vulnerability to developing psychiatric disorders [91, 92]. Future studies carried out on naturalistic cohorts should consider collecting data on the timing but also on the type of stress experienced in early life, to better capture the individual exposome. Finally, DNA methylation is tightly associated with genetic variation [93]. Several studies reporting associations with early life stress or mood disorder symptoms are based on populations of Asian ancestry, whereas our study only included participants of European ancestry. Hence, although we took into account two genetic variants that might influence the expression of SLC6A4 or TPH2, our findings may only be generalizable to populations of European ancestry. In addition, we used the online mQTL database browser (http://www.mqtldb.org/) to investigate potential genetic influences on the methylation loci considered in our study. We did not find any variant associated with any of our CpG sites of interest. However, as only three out of the ten loci were available on their dataset, we cannot exclude that other genetic variants might affect SLC6A4 or TPH2 methylation. Future studies considering more CpG sites and genetic variants as well as including participants from different ancestries would help clarify this limitation.
Conclusions
To conclude, our findings do not support an association between SLC6A4 or TPH2 methylation and 5-HTT or 5-HT4 brain levels or measures of early life stress, anxiety or depressive symptoms. We suggest that caution should be used when interpreting findings on peripheral DNA methylation in relation to the adult serotonergic brain architecture and to measures of early life stress or mood disorders symptoms. However, our findings do not rule out a role of peripheral DNA methylation in serotonergic neurotransmission and (mal)adaptation to environmental stress, which should be further elucidated by future studies considering more CpG sites and related genetic variants, larger sample sizes, more sensitive measures of early environmental stress, blood cell composition and longitudinal cohorts.
Availability of data and materials
The R codes used for statistical analyses can be made available upon request to the corresponding author (vibe.frokjaer@nru.dk). Data can be made available upon reasonable request via this form (https://cimbi.dk/index.php/documents/category/3-cimbi-database) and with an appropriate data sharing agreement.
Abbreviations
- SLC6A4 :
-
Serotonin transporter gene
- TPH2 :
-
Tryptophan hydroxylase 2 gene
- 5-HTT:
-
Serotonin transporter
- 5-HT4 :
-
Serotonin 4 receptor
- MDD:
-
Major depressive disorder
- 5-HTTLPR:
-
Serotonin-transporter-linked promoter region polymorphism
- CpG:
-
Cytosine–Guanine dinucleotide
- HAMD17:
-
Hamilton depression rating scale 17-item
- HAMD6:
-
Hamilton depression rating scale 6-item
- MRTM/MRTM2:
-
Multilinear reference tissue model
- LVM:
-
Latent variable model
- 5-HTTLV :
-
Latent variable based on 5-HTT binding values
- 5-HT4LV :
-
Latent variable based on 5-HT4 binding values
- ROI:
-
Region of interest
- SLE:
-
Stressful life events
- PBI:
-
Parental bonding inventory
- BDI:
-
Beck’s depression index
- CATS:
-
Childhood abuse trauma scale
- GAD10:
-
Generalized anxiety disorder 10-item
- PSS:
-
Perceived stress scale
- CpGLV :
-
Latent variable based on CpG values
- CpGLV+cells :
-
CpGLV adjusted for cell proportions
- HC:
-
Healthy control
References
Lesch KP, Waider J. Serotonin in the modulation of neural plasticity and networks: implications for neurodevelopmental disorders. Neuron. 2012;76(1):175–91. https://doi.org/10.1016/j.neuron.2012.09.013.
Mc Mahon B, Andersen SB, Madsen MK, Hjordt LV, Hageman I, Dam H, et al. Seasonal difference in brain serotonin transporter binding predicts symptom severity in patients with seasonal affective disorder. Brain. 2016;139(5):1605–14.
Portas CM, Bjorvatn B, Ursin R. Serotonin and the sleep/wake cycle: special emphasis on microdialysis studies. Prog Neurobiol. 2000;60(1):13–35.
Štrac DŠ, Pivac N, Mück-Šeler D. The serotonergic system and cognitive function. Transl Neurosci. 2016;7(1):35–49.
Branchi I. The double edged sword of neural plasticity: Increasing serotonin levels leads to both greater vulnerability to depression and improved capacity to recover. Psychoneuroendocrinology. 2011;36(3):339–51. https://doi.org/10.1016/j.psyneuen.2010.08.011.
Carhart-Harris RL, Nutt DJ. Serotonin and brain function: a tale of two receptors. J Psychopharmacol. 2017;31(9):1091–120.
Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, et al. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuro-Psychopharmacology Biol Psychiatry. 2013;44:23–8. https://doi.org/10.1016/j.pnpbp.2013.01.006.
Palma-Gudiel H, Fañanás L. An integrative review of methylation at the serotonin transporter gene and its dialogue with environmental risk factors, psychopathology and 5-HTTLPR. Neurosci Biobehav Rev. 2017;72:190–209.
Walther DJ, Peter JU, Bashammakh S, Hörtnagl H, Voits M, Fink H, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science (80-). 2003;299(5603):76.
Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington HL, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science (80-). 2003;301(5631):386–9.
Culverhouse RC, Saccone NL, Horto AC, et al. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry. 2018;23(1):133–42.
Border R, Johnson EC, Evans LM, Berley N, Sullivan PF, Keller MC. No support for historic candidate gene or candidate gene-by- interaction hypotheses for major depression across multiple large samples. Am J Psychiatry. 2019;176(5):376–87.
Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet Part B Neuropsychiatr Genet. 2007;144(1):101–5.
Vijayendran M, Beach SRH, Plume JM, Brody GH, Philibert RA. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front Psychiatry. 2012;3(June):1–7.
Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimäki M, Vahtera J, et al. Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PLoS ONE. 2012;7(9):3–9.
Leibold NK, Weidner MT, Ziegler C, Ortega G, Domschke K, Lesch KP, et al. DNA methylation in the 5-HTT regulatory region is associated with CO2-induced fear in panic disorder patients. Eur Neuropsychopharmacol. 2020;2020(36):154–9.
Van Ijzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SRH, Philibert R. Methylation matters: Interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry. 2010;68(5):405–7. https://doi.org/10.1016/j.biopsych.2010.05.008.
Okada S, Morinobu S, Fuchikami M, Segawa M, Yokomaku K, Kataoka T, et al. The potential of SLC6A4 gene methylation analysis for the diagnosis and treatment of major depression. J Psychiatr Res. 2014;53(1):47–53. https://doi.org/10.1016/j.jpsychires.2014.02.002.
Zhao J, Goldberg J, Bremner JD, Vaccarino V. Association between promoter methylation of serotonin transporter gene and depressive symptoms: a monozygotic twin study. Psychosom Med. 2013;75(6):523–9.
Schiele MA, Zwanzger P, Schwarte K, Arolt V, Baune BT, Domschke K. Serotonin transporter gene promoter hypomethylation as a predictor of antidepressant treatment response in major depression: a replication study. Int J Neuropsychopharmacol. 2021;24(3):191–9.
Domschke K, Tidow N, Schwarte K, Deckert J, Lesch KP, Arolt V, et al. Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int J Neuropsychopharmacol. 2014;17(8):1167–76.
Moon YK, Kim H, Kim S, Lim SW, Kim DK. Influence of antidepressant treatment on SLC6A4 methylation in Korean patients with major depression. Am J Med Genet Part B Neuropsychiatr Genet. 2023;192(1–2):28–37.
Shen T, Li X, Chen L, Chen Z, Tan T, Hua T, et al. The relationship of tryptophan hydroxylase-2 methylation to early-life stress and its impact on short-term antidepressant treatment response. J Affect Disord [Internet]. 2020;276(April 2020):850–8. https://doi.org/10.1016/j.jad.2020.07.111.
Tan T, Xu Z, Gao C, Shen T, Li L, Chen Z, et al. Influence and interaction of resting state functional magnetic resonance and tryptophan hydroxylase-2 methylation on short-term antidepressant drug response. BMC Psychiatry. 2022;22(1):1–10. https://doi.org/10.1186/s12888-022-03860-z.
Yuan M, Yang B, Rothschild G, Mann JJ, Sanford LD, Tang X, et al. Epigenetic regulation in major depression and other stress-related disorders: molecular mechanisms, clinical relevance and therapeutic potential. Signal Transduct Target Ther. 2023;8(1):309.
Iga J-I, Watanabe S-y, Numata S, Umehara H, Nishi A, Kinoshita M, Inoshita M, Shimodera S, Fujita H, Ohmori T. Association study of polymorphism in the serotonin transporter gene promoter, methylation profiles, and expression in patients with major depressive disorder. Hum Psychopharmacol. 2016;31:193–9. https://doi.org/10.1002/hup.2527.
Wang D, Szyf M, Benkelfat C, Provençal N, Turecki G, Caramaschi D, et al. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS ONE. 2012;7(6):3–10.
Yang X, Shao X, Gao L, Zhang S. Systematic DNA methylation analysis of multiple cell lines reveals common and specific patterns within and across tissues of origin. Hum Mol Genet. 2015;24(15):4374–84.
Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–32.
Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci. 2014;17(9):1153–5. https://doi.org/10.1038/nn.3778.
Riese H, van den Heuvel ER, Snieder H, den Dunnen WF, Plosch T, Kema IP, et al. Association between methylation of the SLC6A4 promoter region in peripheral blood leukocytes and methylation in amygdala tissue. Psychosom Med. 2014;76:244–6.
Haahr ME, Fisher PM, Jensen CG, Frokjaer VG, McMahon B, Madsen K, et al. Central 5-HT4 receptor binding as biomarker of serotonergic tonus in humans: A [11C]SB207145 PET study. Mol Psychiatry. 2014;19(4):427–32.
Houle S, Ginovart N, Hussey D, Meyer JH, Wilson AA. Imaging the serotonin transporter with positron emission tomography: Initial human studies with [11C]DAPP and [11C]DASB. Eur J Nucl Med. 2000;27(11):1719–22.
Marner L, Gillings N, Madsen K, Erritzoe D, Baaré WFC, Svarer C, et al. Brain imaging of serotonin 4 receptors in humans with [11C]SB207145-PET. Neuroimage. 2010;50(3):855–61. https://doi.org/10.1016/j.neuroimage.2010.01.054.
Fisher PMD, Haahr ME, Jensen CG, Frokjaer VG, Siebner HR, Knudsen GM. Fluctuations in [11C]SB207145 PET binding associated with change in threat-related amygdala reactivity in humans. Neuropsychopharmacology. 2015;40(6):1510–8.
Köhler-Forsberg K, Dam VH, Ozenne B, Sankar A, Beliveau V, Landman EB, et al. Serotonin 4 receptor brain binding in major depressive disorder and association with memory dysfunction. JAMA Psychiat. 2023;80(4):296–304.
Larsen SV, Köhler-Forsberg K, Dam VH, Poulsen AS, Svarer C, Jensen PS, et al. Oral contraceptives and the serotonin 4 receptor: a molecular brain imaging study in healthy women. Acta Psychiatr Scand. 2020;142(4):294–306.
Murphy SE, Wright LC, Browning M, Cowen PJ, Harmer CJ. A role for 5-HT4 receptors in human learning and memory. Psychol Med. 2019;50:2722–30.
Moore SR, Kobor MS. Variability in DNA methylation at the serotonin transporter gene promoter: epigenetic mechanism or cell-type artifact? Mol Psychiatry. 2017;25(9):1906–9. https://doi.org/10.1038/s41380-018-0121-6.
Qi L, Teschendorff AE. Cell - type heterogeneity: Why we should adjust for it in epigenome and biomarker studies. Clin Epigenetics. 2022. https://doi.org/10.1186/s13148-022-01253-3.
Wankerl M, Miller R, Kirschbaum C, Hennig J, Stalder T, Alexander N. Effects of genetic and early environmental risk factors for depression on serotonin transporter expression and methylation profiles. Transl Psychiatry. 2014;4(6):e402–9. https://doi.org/10.1038/tp.2014.37.
Knudsen GM, Jensen PS, Erritzoe D, Baaré WFC, Ettrup A, Fisher PM, et al. The center for integrated molecular brain imaging (Cimbi) database. Neuroimage. 2016;124:1213–9. https://doi.org/10.1016/j.neuroimage.2015.04.025.
Köhler-Forsberg K, Jorgensen A, Dam VH, Stenbæk DS, Fisher PM, Ip CT, et al. Predicting treatment outcome in major depressive disorder using serotonin 4 receptor PET brain imaging, functional MRI, cognitive-, EEG-based, and peripheral biomarkers: a neuropharm open label clinical trial protocol. Front Psychiatry. 2020;11(July):1–15.
Dam VH, Stenbæk DS, Köhler-Forsberg K, Ip C, Ozenne B, Sahakian BJ, et al. Evaluating cognitive disturbances as treatment target and predictor of antidepressant action in major depressive disorder: a NeuroPharm study. Transl Psychiatry. 2022;12(1):1–8.
Hamilton M. Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol. 1967;6(4):278–96.
Kim JM, Stewart R, Kang HJ, Kim SW, Shin IS, Kim HR, et al. A longitudinal study of SLC6A4 DNA promoter methylation and poststroke depression. J Psychiatr Res [Internet]. 2013;47(9):1222–7. https://doi.org/10.1016/j.jpsychires.2013.04.010.
Fan R, Hua T, Shen T, Jiao Z, Yue Q, Chen B, et al. Identifying patients with major depressive disorder based on tryptophan hydroxylase-2 methylation using machine learning algorithms. Psychiatry Res. 2021;306(June):114258. https://doi.org/10.1016/j.psychres.2021.114258.
Akhrif A, Romanos M, Peters K, Furtmann AK, Caspers J, Lesch KP, et al. Serotonergic modulation of normal and abnormal brain dynamics: The genetic influence of the TPH2 G-703T genotype and DNA methylation on wavelet variance in children and adolescents with and without ADHD. PLoS ONE. 2023;18(4 April):1–20.
Bruzzone SEP, Nasser A, Aripaka SS, Spies M, Ozenne B, Jensen PS, et al. Genetic contributions to brain serotonin transporter levels in healthy adults: a PET study. Sci Rep. 2023. https://doi.org/10.1038/s41598-023-43690-x.
Fisher PM, Ozenne B, Svarer C, Adamsen D, Lehel S, Baaré WFC, et al. BDNF val66met association with serotonin transporter binding in healthy humans. Transl Psychiatry. 2017;7(2):e1029–36. https://doi.org/10.1038/tp.2016.295.
Gutknecht L, Jacob C, Strobel A, Kriegebaum C, Müller J, Zeng Y, et al. Tryptophan hydroxylase-2 gene variation influences personality traits and disorders related to emotional dysregulation. Int J Neuropsychopharmacol. 2007;10(3):309–20.
Köhler-Forsberg K, Dam VH, Ozenne B, Sankar A, Beliveau V, Landman EB, et al. Serotonin 4 receptor brain binding in major depressive disorder and association with memory dysfunction. JAMA Psychiatry [Internet]. 2023;1–9. https://jamanetwork.com/journals/jamapsychiatry/fullarticle/2801424
Svarer C, Madsen K, Hasselbalch SG, Pinborg LH, Haugbøl S, Frøkjær VG, et al. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage. 2005;24(4):969–79.
R Core Team. R: A Language and environment for statistical computing. Vienna, Austria; 2021. https://www.r-project.org/
Beliveau V, Ganz M, Feng L, Ozenne B, Højgaard L, Fisher PM, et al. A high-resolution in vivo atlas of the human brain’s serotonin system. J Neurosci. 2017;37(1):120–8.
Madsen K, Torstensen E, Holst KK, Haahr ME, Knorr U, Frokjaer VG, et al. Familial risk for major depression is associated with lower striatal 5-HT4 receptor binding. Int J Neuropsychopharmacol. 2015;18(1):1–7.
Holst KK, Budtz-Jørgensen E. Linear latent variable models: The lava-package. Comput Stat. 2013;28:1385–452.
Fisher PM, Holst KK, Adamsen D, Klein AB, Frokjaer VG, Jensen PS, et al. BDNF Val66met and 5-HTTLPR polymorphisms predict a human in vivo marker for brain serotonin levels. Hum Brain Mapp. 2015;36(1):313–23.
Madsen K, Haahr MT, Marner L, Keller SH, Baaré WF, Svarer C, et al. Age and sex effects on 5-HT 4 receptors in the human brain: A 11 CSB207145 PET study. J Cereb Blood Flow Metab. 2011;31(6):1475–81.
Yamamoto M, Suhara T, Okubo Y, Ichimiya T, Sudo Y, Inoue M, et al. Age-related decline of serotonin transporters in living human brain of healthy males. Life Sci. 2002;71(7):751–7.
Aberg KA, Dean B, Shabalin AA, Chan RF, Han LKM, Zhao M, et al. Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples. Mol Psychiatry. 2020;25(6):1344–54. https://doi.org/10.1038/s41380-018-0247-6.
Chatterjee R, Vinson C. CpG methylation recruits sequence specific transcription factors essential for tissue specific gene expression. Biochim Biophys Acta - Gene Regul Mech. 2012;1819(7):763–70. https://doi.org/10.1016/j.bbagrm.2012.02.014.
Zhang Y, Chang Z, Chen J, Ling Y, Liu X, Feng Z, et al. Methylation of the tryptophan hydroxylase-2 gene is associated with mRNA expression in patients with major depression with suicide attempts. Molecular Med Rep. 2015;12:3184–90.
Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. MiR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science (80-). 2010;329(5998):1537–41.
Bradley CC, Blakely RD. Alternative splicing of the human serotonin transporter gene. J Neurochem. 1997;69(4):1356–67.
Millan MJ. MicroRNA in the regulation and expression of serotonergic transmission in the brain and other tissues. Curr Opin Pharmacol [Internet]. 2011;11(1):11–22. https://doi.org/10.1016/j.coph.2011.01.008.
Grohmann M, Hammer P, Walther M, Paulmann N, Bu A, Rupprecht R, et al. Alternative splicing and extensive RNA editing of human TPH2 transcripts. 2010;5(1).
Spies M, Murga M, Vraka C, Philippe C, Gryglewski G, Nics L, et al. Impact of genetic variants within serotonin turnover enzymes on human cerebral monoamine oxidase A in vivo. 2023;(June).
Pratelli M, Pasqualetti M. Serotonergic neurotransmission manipulation for the understanding of brain development and function: learning from Tph2 genetic models. Biochimie [Internet]. 2019;161:3–14. https://doi.org/10.1016/j.biochi.2018.11.016.
Witteveen JS, Middelman A, van Hulten JA, Martens GJM, Homberg JR, Kolk SM. Lack of serotonin reuptake during brain development alters rostral raphe-prefrontal network formation. Front Cell Neurosci. 2013;7(Oct):1–16.
Jakobsen GR, Fisher PM, Dyssegaard A, McMahon B, Holst KK, Lehel S, et al. Brain serotonin 4 receptor binding is associated with the cortisol awakening response. Psychoneuroendocrinology. 2016;67:124–32.
Kim DK, Tolliver TJ, Huang SJ, Martin BJ, Andrews AM, Wichems C, et al. Altered serotonin synthesis, turnover and dynamic regulation in multiple brain regions of mice lacking the serotonin transporter. Neuropharmacology. 2005;49(6):798–810.
Praschak-Rieder N, Willeit M, Wilson AA, Houle S, Meyer JH. Seasonal variation in human brain serotonin transporter binding. Arch Gen Psychiatry. 2008;65(9):1072–8.
Vulpius GM, Köhler-forsberg K, Ozenne B, Larsen SV, Nasser A, Svarer C, et al. Stress hormone dynamics are coupled to brain serotonin 4 receptor availability in unmedicated patients with major depressive disorder: a neuropharm study. Int J Neuropsychopharmacol. 2023;26(August):639–48.
Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–86.
Mulder RH, Neumann A, Cecil CAM, Walton E, Houtepen LC, Simpkin AJ, et al. Epigenome-wide change and variation in DNA methylation in childhood: trajectories from birth to late adolescence. Hum Mol Genet. 2021;30(1):119–34.
Elwenspoek MMC, Kuehn A, Muller CP, Turner JD. The effects of early life adversity on the immune system. Psychoneuroendocrinology. 2017;82:140–54. https://doi.org/10.1016/j.psyneuen.2017.05.012.
Hunt C, Macedo e Cordeiro T, Suchting R, de Dios C, Cuellar Leal VA, Soares JC, et al. Effect of immune activation on the kynurenine pathway and depression symptoms: a systematic review and meta-analysis. Neurosci Biobehav Rev. 2020;118:514–23. https://doi.org/10.1016/j.neubiorev.2020.08.010.
Shen T, Li X, Chen L, Chen Z, Tan T, Hua T, et al. The relationship of tryptophan hydroxylase-2 methylation to early-life stress and its impact on short-term antidepressant treatment response. J Affect Disord. 2020;276(April):850–8. https://doi.org/10.1016/j.jad.2020.07.111.
Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults : a systematic review of DNA methylation studies. Clin Epigenetics. 2015. https://doi.org/10.1186/s13148-015-0148-3.
Liu C, Marioni RE, Hedman ÅK, Pfeiffer L, Tsai P, Reynolds LM, et al. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry. 2018;23:422–33.
Rider CF, Carlsten C. Air pollution and DNA methylation: effects of exposure in humans. 2019;1–15.
Köhler-Forsberg K, Dam VH, Ozenne B, Sankar A, Beliveau V, Landman EB, et al. Serotonin 4 receptor brain binding in major depressive disorder and association with memory dysfunction. JAMA Psychiatry. 2023;1–9.
Philibert, Robert A., Sandhu, Harinder, Hollenbeck, Nancy, Gunter, Tracy, Adams, William, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;543–549.
Koestler DC, Christensen BC, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8(8):816–26.
Gryglewski G, Lanzenberger R, Kranz GS, Cumming P. Meta-analysis of molecular imaging of serotonin transporters in major depression. J Cereb Blood Flow Metab. 2014;34(7):1096–103.
Jennings KA, Licht CL, Bruce A, Lesch KP, Knudsen GM, Sharp T. Genetic variation in 5-hydroxytryptamine transporter expression causes adaptive changes in 5-HT4 receptor levels. Int J Neuropsychopharmacol. 2012;15(8):1099–107.
Rebholz H, Friedman E, Castello J. Alterations of expression of the serotonin 5-HT4 receptor in brain disorders. Int J Mol Sci. 2018;19(11):3581.
Provençal N, Binder EB. The effects of early life stress on the epigenome: From the womb to adulthood and even before. Exp Neurol. 2015;268:10–20. https://doi.org/10.1016/j.expneurol.2014.09.001.
Burns SB, Szyszkowicz JK, Luheshi GN, Lutz PE, Turecki G. Plasticity of the epigenome during early-life stress. Semin Cell Dev Biol. 2018;77:115–32. https://doi.org/10.1016/j.semcdb.2017.09.033.
Herbison CE, Allen K, Robinson M, Newnham J. The impact of life stress on adult depression and anxiety is dependent on gender and timing of exposure. Dev Psychopathol. 2017;29:1443–54.
Colli CD, Borgi M, Poggini S, Chiarotti F, Cirulli F. Time moderates the interplay between 5-HTTLPR and stress on depression risk : gene x environment interaction as a dynamic process. Transl Psychiatry. 2022;12(June):1–12.
Villicaña S, Bell JT. Genetic impacts on DNA methylation: research findings and future perspectives. Genome Biol. 2021;22(1):1–35.
Acknowledgements
We would like to thank A. Nasser and S. Sagar Aripaka for the technical support provided for DNA extraction and genotyping, and N. Schraud for her help with genotyping.
We thank all the participants who agreed on taking part into the studies that allowed to create the dataset used in this article. We also thank all the students, researchers and technical staff that helped collecting the data used in this study. In particular, we acknowledge L. Freyr, S. V. Larsen, G. Thomsen, S. Olsen for their help with the data collection with both the PET and MR scanners. We also acknowledge the John and Birthe Meyer Foundation for donating the PET scanner and the Cyclotron.
Funding
The work described in this study was supported by the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. 953327 (Serotonin and Beyond), Ivan Nielsen foundation and the Læge Sofus Carl Emil Friis og Hustru Olga Doris Friis' Legat. The Center for Integrated Molecular Brain Imaging (https://doi.org/10.1016/j.neuroimage.2015.04.025) was supported by the Danish National Research Council, the Research Council of Rigshospitalet (Grant ID A6594) and the Lundbeck Foundation (GrantID: R279-2018–1145). The NeuroPharm-NP1 study was supported by the Innovation Fund Denmark (GrantID: 5189-00087A).
Author information
Authors and Affiliations
Contributions
SEPB took care of conceptualization, DNA methylation measurements, genotyping, data analysis, writing of the original draft, design and execution of figures and tables, manuscript editing and reviewing and funding. BO supported conceptualization, statistical analyses, manuscript editing and reviewing. PMF helped in conceptualization, data analysis, manuscript editing and reviewing and supervision. GO contributed to the DNA methylation measurements and genotyping. PSJ helped with data management, manuscript editing and reviewing. VHD was involved in data collection, data management, manuscript editing and reviewing. CS contributed to PET data analysis, manuscript editing and reviewing. GMK provided funding, manuscript editing and reviewing and supervision. KPL supplied supervision, funding, manuscript editing and reviewing. VGF contributed to conceptualization, funding, clinical care of MDD patients, manuscript editing and reviewing and supervision. All co-authors approved the final draft of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All studies protocols comply with the Declaration of Helsinki. All participants provided written and informed consent before being included in the research studies. Data of healthy participants included in this study are from the Cimbi database (https://doi.org/10.1016/j.neuroimage.2015.04.025). All research projects generating the data included in the Cimbi database have been approved by the Danish Data Protection Agency and the local scientific ethics committee. Data of patients with major depressive disorder included in this study are from the NeuroPharm 1 study, a clinical trial registered at clinicaltrials.gov (NCT02869035) on 16 August 2016. The study protocol was approved by the Health Research Ethics Committees of the Capital Region of Denmark (reference number: H-15017713), the Danish Medicines Agency (protocol number: NeuroPharm-NP1, EudraCT-number 2016–001626-34) and the Danish Data Protection Agency (04711/RH-2016–163).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
13148_2024_1678_MOESM6_ESM.xlsx
Table S6. Associations between SLC6A4 methylation and environmental stress in healthy controls (HC) and mood and anxiety disorder symptoms in patients with depression (MDD) (XLSX 11 kb)
13148_2024_1678_MOESM7_ESM.xlsx
Table S7. Associations between TPH2 methylation and environmental stress in healthy controls (HC) and mood and anxiety disorder symptoms in patients with depression (MDD) (XLSX 12 kb)
13148_2024_1678_MOESM9_ESM.xlsx
Table S9. Results of associations between CpGLV+cells and measures of environmental stress or mood or anxiety symptoms (XLSX 10 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bruzzone, S.E.P., Ozenne, B., Fisher, P.M. et al. No association between peripheral serotonin-gene-related DNA methylation and brain serotonin neurotransmission in the healthy and depressed state. Clin Epigenet 16, 71 (2024). https://doi.org/10.1186/s13148-024-01678-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-024-01678-y