Abstract
Background
Multiple studies have reported the prognostic impact of DNA methylation changes in acute myeloid leukemia (AML). However, these epigenetic markers have not been thoroughly validated and therefore are still not considered in clinical practice. Hence, we aimed to independently verify results of selected studies describing the relationship between DNA methylation of specific genes and their prognostic potential in predicting overall survival (OS) and event-free survival (EFS).
Results
Fourteen studies (published 2011–2019) comprising of 27 genes were subjected to validation by a custom NGS-based sequencing panel in 178 newly diagnosed non-M3 AML patients treated by 3 + 7 induction regimen. The results were considered as successfully validated, if both the log-rank test and multivariate Cox regression analysis had a p-value ≤ 0.05. The predictive role of DNA methylation was confirmed for three studies comprising of four genes: CEBPA (OS: p = 0.02; EFS: p = 0.03), PBX3 (EFS: p = 0.01), LZTS2 (OS: p = 0.05; EFS: p = 0.0003), and NR6A1 (OS: p = 0.004; EFS: p = 0.0003). For all of these genes, higher methylation was an indicator of longer survival. Concurrent higher methylation of both LZTS2 and NR6A1 was highly significant for survival in cytogenetically normal (CN) AML group (OS: p < 0.0001; EFS: p < 0.0001) as well as for the whole AML cohort (OS: p = 0.01; EFS < 0.0001). In contrast, for two studies reporting the poor prognostic effect of higher GPX3 and DLX4 methylation, we found the exact opposite, again linking higher GPX3 (OS: p = 0.006; EFS: p < 0.0001) and DLX4 (OS: p = 0.03; EFS = 0.03) methylation to a favorable treatment outcome. Individual gene significance levels refer to the outcomes of multivariate Cox regression analysis.
Conclusions
Out of twenty-seven genes subjected to DNA methylation validation, a prognostic role was observed for six genes. Therefore, independent validation studies are necessary to reveal truly prognostic DNA methylation changes and to enable the introduction of these promising epigenetic markers into clinical practice.
Similar content being viewed by others
Introduction
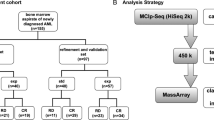
Acute myeloid leukemia (AML) is a hematopoietic malignancy characterized by a complex interplay of aberrations at different levels of the genome (i.e., genetic, epigenetic, transcriptomic, and proteomic) [1,2,3]. This complexity is faithfully reflected by AML heterogeneity in terms of pathogenesis and prognosis. In clinical practice, only properly introduced and validated genetic lesions altogether with cytogenetics are considered into treatment decision making [4]. This still applies despite growing evidence that some other markers, such as epigenetic factors, may add valuable information about the predicted course of the disease in individual AML patients [3]. DNA methylation is one of the longest-studied epigenetic mechanisms and is stable and relatively easy to measure [5, 6]. Therefore, its status can be readily harnessed as a clinically relevant stratifier. Indeed, there are an increasing number of articles assessing the influence of DNA methylation on AML prognosis—reviewed in [7]. These studies interrogate one, a few or multiple loci depending on the methodology used. Typically, as a result of such research, authors define gene(s) that may serve as new biomarkers to improve risk stratification in AML patients. The main weakness is that such works are usually not validated by other researchers and hence there is not sufficient validation of these potential biomarkers for them to be introduced into clinical practice. Therefore, we designed a comprehensive NGS-based DNA methylation panel comprising of genes previously published as having an impact on AML prognosis. For validation purposes, we selected fourteen studies published between years 2011 and 2019 [8,9,10,11,12,13,14,15,16,17,18,19,20,21] covering 27 genes (Additional file 1: Table S1). We chose works targeting only one or a few loci at once (averaged 2 loci per publication, range 1 to 7), because lower numbers of biomarkers would be more feasible for introduction into a laboratory routine practice. The list of the selected studies and their basic characterization is summarized in Table 1. The aim of this work was to make an independent verification of results published by other researchers to narrow down the list of actually prognostically relevant genes that may allow more precise AML stratification in the future.
Results
Our validation study confirmed association of DNA methylation status and prognosis for four genes: CEBPA [13], PBX3 [10], UZTS2 [16], and NR6A1 [16]. A summary of the results is presented in Table 2. Surprisingly, for two studies [19, 20], we found the exact opposite effect of DNA methylation on prognosis than originally reported—higher GPX3 and DLX4 methylation—was linked to a better outcome according to our data. Kaplan–Meier curves for OS and EFS for all six significant genes are shown in Figs. 1 and 2, respectively. In four additional studies [8, 9, 15, 21], only the results from log-rank test displayed statistical significance that was lost in the subsequent multivariate testing (Table 2). These results were not considered as sufficiently conclusive for classifying them as validated. The mean DNA methylation values in hypo- versus hypermethylated subgroups for each of the significant genes are depicted in Fig. 3.
Kaplan–Meier (KM) curves for overall survival (OS): A CEBPA methylation KM curves in AML subgroup excluding favorable cytogenetics and without CEBPA and NPM1 mutations (n = 83). B GPX3 methylation KM curves in the whole non-M3 AML cohort (n = 178). C DLX4 methylation KM curves in the whole non-M3 AML cohort (n = 178). D LZTS2 methylation KM curves in the whole non-M3 AML cohort (n = 178). E NR6A1 methylation KM curves in the whole non-M3 AML cohort (n = 178). F LZTS2&NR6A1 methylation KM curves in the whole non-M3 AML cohort (n = 178). G LZTS2 methylation KM curves in the CN-AML subgroup (n = 85). H NR6A1 methylation KM curves in the CN-AML subgroup (n = 85). I LZTS2&NR6A1 methylation KM curves in the CN-AML subgroup (n = 85). CN-AML = cytogenetically normal AML, hypo = hypomethylated, hyper = hypermethylated, Strata—stratified by a variable
Kaplan–Meier (KM) curves for event-free survival (EFS): A CEBPA methylation KM curves in AML subgroup excluding favorable cytogenetics and without CEBPA and NPM1 mutations (n = 83). B PBX3 methylation KM curves in the whole non-M3 AML cohort (n = 178). C GPX3 methylation KM curves in the whole non-M3 AML cohort (n = 178). D DLX4 methylation KM curves in the whole non-M3 AML cohort (n = 178). E LZTS2 methylation KM curves in the whole non-M3 AML cohort (n = 178). F NR6A1 methylation KM curves in the whole non-M3 AML cohort (n = 178). G LZTS2&NR6A1 methylation KM curves in the whole non-M3 AML cohort (n = 178). H LZTS2 methylation KM curves in the CN-AML subgroup (n = 85). I NR6A1 methylation KM curves in the CN-AML subgroup (n = 85). J LZTS2&NR6A1 methylation KM curves in the CN-AML subgroup (n = 85). CN-AML = cytogenetically normal AML, hypo = hypomethylated, hyper = hypermethylated, Strata—stratified by a variable
Discussion
Despite a large number of studies addressing the importance of DNA methylation changes for AML prognosis, these aberrations are still not considered for risk stratification, although many promising results have been already reported. The lack of independent validation studies is probably the main obstacle that does not allow the implementation of epigenetic markers alongside the well-established genetic ones. Most of the publications present just more new potential epigenetic biomarkers, making the actual role of DNA methylation harder to grasp and interpret for clinical purposes. With the aim to verify the prognostic role of specific and already described DNA methylation changes in AML, we designed our custom NGS-based DNA methylation panel that covers 27 genes (Additional file 1: Table S1) taken from 14 studies published between years 2011 and 2019. The reported prognostic significance was verified for three studies [10, 13, 16]. These three studies do not share any apparent features such as size of test cohort, presence of a validation cohort, methodology, or biological material utilized for the DNA methylation assessment (see Table 1). We briefly summarize and discuss the genes with a confirmed role of DNA methylation in AML prognosis. CEBPA is a well-known gene involved in AML pathogenesis. Double CEBPA mutations have been connected to better OS and EFS [4]. Con cordantly, hypermethylation of distal CEBPA promoter was reported as a favorable prognostic biomarker, which we proved in AML subgroup excluding favorable cytogenetics and without CEBPA and NPM1 mutations, but not in CN-AML without CEBPA and NPM1 mutations as also originally described by Lin et al. [13]. PBX3 has been identified as an oncogene in AML that transcriptionally regulates HOXA genes and promotes cell proliferation and resistance to chemotherapeutical agents [22]. Hajkova et al. [10] reported PBX3 overexpression associated with a higher incidence of relapses. They also showed a clear correlation between PBX3 overexpression and hypomethylation. In line with this, we detected PBX3 hypomethylation as an independent negative prognostic factor for EFS. Qu et al. [16] identified higher methylation in CpG island (CGI) shores of LZTS2 and NR6A1 genes as a predictor of better prognosis in CN-AML. Interestingly, we confirmed the predictive role of LZTS2 and NR6A1 hypermethylation not only in CN-AML, but in the whole non-M3 diagnostic AML cohort as well. The strongest link between DNA methylation and prognosis was observed if the concurrent hypermethylation of both genes was present. Validation of the works of Zhou et al. [19, 20] produced contradictory results to the original studies. Unlike them, we observed a clear association between higher GPX3/DLX4 promoter methylation and better survival. This discrepancy is hard to explain because even usage of different methodology (qMSP versus NGS) or biological material (BM versus PB) would not completely reverse the impact of particular gene’s hypermethylation. The recent GPX3 review described its dichotomous role in different cancer types; it can act as either an oncogene or a tumor suppressor [23]. Tumors with high GPX3 expression have an increased resistance to chemotherapy due to the GPX3 involvement in the antioxidant enzyme system [24]. This would support our findings about GPX3 hypermethylation (and thus probable downregulation) and favorable outcome in AML cohort treated by standard 3 + 7 induction regimen. As for DLX4, its overexpression was described in numerous tumor types (including AML) in association with tumor progression and/or invasion [25,26,27,28]. This again supports the link between DLX4 hypermethylation and better AML prognosis.
Noticeably, all verified prognostic DNA methylation changes have one thing in common: higher methylation equals better prognosis. Six out of fourteen studies subjected to the validation reported higher methylation/lower expression and superior outcome. From these six studies, three were verified by both log-rank and multivariate Cox regression analysis [10, 13, 16] and three showed significance by log-rank test [8, 15, 21]. On the other hand, from eight studies describing the relationship between higher methylation and poor prognosis, only one displayed significance by log-rank test [9], none was verified by multivariate Cox regression analysis, and for two studies the opposite relation between higher methylation and prognosis was revealed [19, 20]. Altogether, it seems that higher methylation has predominant influence on prognosis in AML. However, the exact location of differential methylation and what specific genes are affected are probably the key elements determining the direction of how DNA methylation influences patients’ outcome.
In three studies, the indirect relation of DNA methylation (through its association with gene expression) and prognosis was reported [10, 12, 15]. From these, only one study was validated [10]. Technically speaking, we cannot exclude the role of gene expression deregulation in patients’ outcome in the remaining two studies [12, 15], because in our study design we did not examine the impact of gene expression on AML prognosis.
Another important aspect to discuss is the usage of PB versus BM for DNA methylation assessment. Our AML cohort consists of PB samples only, whereas PB alone was a starting material in 3/14 studies that underwent validation. Some articles have already dealt with the comparison of DNA methylation results obtained from PB versus BM, and they reported their interchangeability for these purposes [8, 10, 16]. In line with this, the result of DNA methylation validation was not determined by the biological material used. In fact, genes with validated role of their methylation status in AML prognosis were all revealed in studies using either BM alone [13, 19, 20] or studies using a combination of PB and BM [10, 16]. PB is a starting material that is easily accessible to the majority of laboratories and it is not as burdensome for patients as BM aspirates.
In practical terms, implementation of a new biomarker represented by a single gene/region is always more feasible than that of a complex methylation pattern. The low number of genes for which we confirmed the prognostic impact with our NGS-based approach highlights the importanc e of such validation and a need for a consistent and easily reproducible approach to assess the impact of various changes in DNA methylation on AML prognosis.
Conclusions
We showed that validation of previously published prognostically significant DNA methylation changes is essential to confirm their relevance for patients’ stratification. Out of 27 genes, a statistically significant correlation between DNA methylation status and prognosis was proved for six of them: CEBPA, PBX3, LZTS2, NR6A1, GPX3, and DLX4. We propose that further independent validation studies may build upon our results, because only markers properly verified by several independent studies can be considered for AML prognosis refinement in clinical practice.
Methods
Patients
We examined 178 adult AML patients: 128 patients from the Institute of Hematology and Blood Transfusion (Prague, Czech Republic) and 50 patients from the University Hospital Brno (Brno, Czech Republic). All patients were diagnosed with AML between 2013 and 2016 and were treated with curative intent starting with 3 + 7 induction regimen [29]. The clinical and basic molecular characteristics used for statistical analysis are stated in Additional file 1: Table S2. Healthy donors (n = 11) were also analyzed. The study was approved by the Ethics committees of both participating institutions and all patients provided their full consent. The research conforms to The Code of Ethics of the World Medical Assoc iation.
Targeted bisulfite sequencing
Sequencing libraries consisted of 16–18 samples and were prepared according to the SeqCap Epi protocol (Roche, Basel, Switzerland) with KAPA HyperPrep Kit (Roche). Diagnostic whole-blood DNA from AML patients (800–1200 ng) was first mixed with the Bisulfite-conversion Control (unmethylated DNA from phage lambda) provided in the SeqCap Epi Accessory kit (Roche) and then fragmented either via E220 Focused ultrasonicator (Covaris, Woburn, MA, USA) or Bioruptor Pico instrument (Diagenode, Liège, Belgium) to get an average size of 200 bp. EZ DNA Methylation Lightning Kit (Zymo Research, Irvine, CA, USA) was used for the bisulfite conversion. Pooled samples from each library were hybridized for about 68 h with a custom set of probes (made by Roche Company). The final concentration of the libraries was measured using KAPA Library Quantification Kit (Roche), and the average size of the libraries’ fragments was assessed on 4200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA). Libraries were sequenced on a MiSeq instrument (Illumina, San Diego, CA, USA) using the MiSeq Reagent Kit v2 (300-cycles) (Illumina).
Sequencing data analysis
FastQC (version 0.11.8) [30] and MultiQC (version 1.7) [31] software was used to check the quality of fastq files. Reads were then trimmed and filtered using Cutadapt (version 2.4) [32] and the quality of reads was checked again. Filtered data were mapped with software Segemehl (version 0.3.4) [33] to human genome version GRCh37/hg19 with added sequence of Enterobacteria phage lambda NC_001416.1. Mapping statistics were assessed and we checked that more than 80% of reads were mapped for each sample. Bam files containing mapped reads were sorted and indexed by Samtools software (version 1.10). Subsequently, we used Haarz tool (version 0.3.4) [33] with enabled "callmethyl" option to select methylated positions and create vcf files that were further processed in R software. Positions that corresponded to the lambda phage sequence were separated and used to check that the bisulfite conversion ratio was > 99% for each sample. Remaining positions were filtered and only CpG positions were left in the data. Finally, we selected regions corresponding to loci published in the original articles results and the average methylation across the regions was assessed. The list of selected regions is provided in Additional file 1: Table S1. Raw sequencing data are available at the Gene Expression Omnibus repository (accession number GSE165435).
Statistical analyses and definitions
For the statistical analyses, R software (version 4.0.0) was used. Surviving patients were censored to the April 6, 2020. Overall survival (OS) was established as time from diagnosis until death of any cause. Event-free survival (EFS) was established as time from the first complete remission until death or hematological relapse. Multivariate Cox regression analysis was computed with following covariates: age, leukocyte count, cytogenetics [34], transplantation in the first complete remission, presence of FLT3-ITD and NPM1 mutations. For five studies (see Table 2), Cutoff Finder [35] was utilized to determine the optimal DNA methylation threshold. We used the same DNA methylation threshold as originally published or it was set up in the most similar and meaningful way. We also adapted the selection of AML patients because some studies detected a prognostic effect of DNA methylation only in a specific subset of AML such as cytogenetically normal (CN) AML. To properly evaluate the prognostic significance of the studied regions, we performed Kaplan–Meier analysis with log-rank test. Subsequently, we assessed the effect of DNA methylation levels on overall (OS) and event-free survival (EFS) using multivariate Cox regression for those loci significantly affecting OS or EFS in Kaplan–Meier analysis. p-value ≤ 0.05 was considered as statistically significant.
Availability of data and materials
Raw DNA methylation sequencing data are deposited into GEO repository with the accession number GSE165435 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165435). The other data supporting the findings of the present study are included in this published article [and its Additional files].
References
Handschuh L. Not only mutations matter: molecular picture of acute myeloid leukemia emerging from transcriptome studies. J Oncol. 2019;30(2019):7239206.
Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118(4):1069–76.
Jiang H, Ou Z, He Y, Yu M, Wu S, Li G, et al. DNA methylation markers in the diagnosis and prognosis of common leukemias. Signal Transduct Target Ther. 2020;5(1):1–10.
Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–47.
Sestakova S, Salek C, Remesova H. DNA methylation validation methods: a coherent review with practical comparison. Biol Proced. 2019;21:1–11.
Gosselt HR, Griffioen PH, van Zelst BD, Oosterom N, de Jonge R, Heil SG. Global DNA (hydroxy)methylation is stable over time under several storage conditions and temperatures. Epigenetics. 2021;16(1):45–53.
Yang X, Wong MPM, Ng RK. Aberrant DNA methylation in acute myeloid leukemia and its clinical implications. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms20184576.
Hajkova H, Markova J, Haskovec C, Sarova I, Fuchs O, Kostecka A, et al. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk Res. 2012;36(9):1128–33.
Guo H, Zhang TJ, Wen XM, Zhou JD, Ma JC, An C, et al. Hypermethylation of secreted frizzled-related proteins predicts poor prognosis in non-M3 acute myeloid leukemia. Onco Targets Ther. 2017;20(10):3635–44.
Hajkova H, Fritz MH, Haskovec C, Schwarz J, Salek C, Markova J, et al. CBFB-MYH11 hypomethylation signature and PBX3 differential methylation revealed by targeted bisulfite sequencing in patients. J Hematol Oncol. 2014;7:66014–664.
Jost E, Lin Q, Weidner CI, Wilop S, Hoffmann M, Walenda T, et al. Epimutations mimic genomic mutations of DNMT3A in acute myeloid leukemia. Leukemia. 2014;28(6):1227–34.
Li XX, Zhou JD, Zhang TJ, Yang L, Wen XM, Ma JC, et al. Epigenetic dysregulation of NKD2 is a valuable predictor assessing treatment outcome in acute myeloid leukemia. J Cancer. 2017;8(3):460–8.
Lin TC, Hou HA, Chou WC, Ou DL, Yu SL, Tien HF, et al. CEBPA methylation as a prognostic biomarker in patients with de novo acute myeloid leukemia. Leukemia. 2011;25(1):32–40.
Liu F, Gong M, Gao L, Cai X, Zhang H, Ma Y. RASSF1A hypermethylation is associated with ASXL1 mutation and indicates an adverse outcome in non-M3 acute myeloid leukemia. Onco Targets Ther. 2017;22(10):4143–51.
Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32(6):548–56.
Qu X, Othus M, Davison J, Wu Y, Yan L, Meshinchi S, et al. Prognostic methylation markers for overall survival in cytogenetically normal patients with acute myeloid leukemia treated on SWOG trials. Cancer. 2017;123(13):2472–81.
Sestakova S, Krejcik Z, Folta A, Cerovska E, Salek C, Merkerova MD, et al. DNA methylation and hydroxymethylation patterns in acute myeloid leukemia patients with mutations in DNMT3A and IDH1/2 and their combinations. Cancer Biomark. 2019;25(1):43–51.
Treppendahl MB, Qiu X, Sogaard A, Yang X, Nandrup-Bus C, Hother C, et al. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood. 2012;119(1):206–16.
Zhou JD, Yao DM, Zhang YY, Ma JC, Wen XM, Yang J, et al. GPX3 hypermethylation serves as an independent prognostic biomarker in non-M3 acute myeloid leukemia. Am J Cancer Res. 2015;5(5):1786–94.
Zhou JD, Zhang TJ, Wang YX, Yang DQ, Yang L, Ma JC, et al. DLX4 hypermethylation is a prognostically adverse indicator in de novo acute myeloid leukemia. Tumour Biol. 2016;37(7):8951–60.
Bozic T, Lin Q, Frobel J, Wilop S, Hoffmann M, Muller-Tidow C, et al. DNA-methylation in C1R is a prognostic biomarker for acute myeloid leukemia. Clin Epigenetics. 2015;7:116015–201536.
Morgan R, Pandha HS. PBX3 in Cancer. Cancers (Basel). 2020;12:2. https://doi.org/10.3390/cancers12020431.
Chang C, Worley BL, Phaeton R, Hempel N. Extracellular glutathione peroxidase GPx3 and its role in cancer. Cancers (Basel). 2020;12:8. https://doi.org/10.3390/cancers12082197.
Pelosof L, Yerram S, Armstrong T, Chu N, Danilova L, Yanagisawa B, et al. GPX3 promoter methylation predicts platinum sensitivity in colorectal cancer. Epigenetics. 2017;12(7):540–50.
Sun G, Ge Y, Zhang Y, Yan L, Wu X, Ouyang W, et al. Transcription factors BARX1 and DLX4 contribute to progression of clear cell renal cell carcinoma via promoting proliferation and epithelial-mesenchymal transition. Front Mol Biosci. 2021;8:626328.
Zhang L, Wan Y, Jiang Y, Zhang Z, Shu S, Cheng W, et al. Overexpression of BP1, an isoform of homeobox gene DLX4, promotes cell proliferation, migration and predicts poor prognosis in endometrial cancer. Gene. 2019;30(707):216–23.
Zhou JD, Yang J, Guo H, Deng ZQ, Wen XM, Yang L, et al. BP1 overexpression is associated with adverse prognosis in de novo acute myeloid leukemia. Leuk Lymphoma. 2016;57(4):828–34.
Hollington P, Neufing P, Kalionis B, Waring P, Bentel J, Wattchow D, et al. Expression and localization of homeodomain proteins DLX4, HB9 and HB24 in malignant and benign human colorectal tissues. Anticancer Res. 2004;24(2B):955–62.
Ferrara F, Vitagliano O. Induction therapy in acute myeloid leukemia: is it time to put aside standard 3 + 7? Hematol Oncol. 2019;37(5):558–63.
Andrews S. FastQC: a quality control tool for high throughput sequence data [Online]. 2015. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 20 June 2020.
Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–8.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–2.
Otto C, Stadler PF, Hoffmann S. Fast and sensitive mapping of bisulfite-treated sequencing data. Bioinformatics. 2012;28(13):1698–704.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–65.
Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE. 2012;7(12):e51862.
Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013 May 30;368(22):2059-2074.
Acknowledgements
Not applicable.
Funding
This study was supported by the Ministry of Health of the Czech Republic, project for conceptual development of research organizations (00023736, IHBT).
Author information
Authors and Affiliations
Contributions
SS and EC performed the experiments. SS, DK, and HR analyzed data. SS, EC, CS, IJ, AF, JM, ZR, PC, and HR interpreted the data. SS, CS, and HR wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed consent was obtained from all patients. The research was approved by the Ethics Committees of the Institute of Hematology and Blood Transfusion (Prague, Czech Republic) and the University Hospital Brno (Brno, Czech Republic).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
List of analyzed regions (positions according to hg19 assembly).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Šestáková, Š., Cerovská, E., Šálek, C. et al. A validation study of potential prognostic DNA methylation biomarkers in patients with acute myeloid leukemia using a custom DNA methylation sequencing panel. Clin Epigenet 14, 22 (2022). https://doi.org/10.1186/s13148-022-01242-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-022-01242-6