Abstract
Objective
In past work in budding yeast, we identified a nucleosomal region required for proper interactions between the histone chaperone complex yFACT and transcribed genes. Specific histone mutations within this region cause a shift in yFACT occupancy towards the 3’ end of genes, a defect that we have attributed to impaired yFACT dissociation from DNA following transcription. In this work we wished to assess the contributions of DNA sequences at the 3’ end of genes in promoting yFACT dissociation upon transcription termination.
Results
We generated fourteen different alleles of the constitutively expressed yeast gene PMA1, each lacking a distinct DNA fragment across its 3’ end, and assessed their effects on occupancy of the yFACT component Spt16. Whereas most of these alleles conferred no defects on Spt16 occupancy, one did cause a modest increase in Spt16 binding at the gene’s 3’ end. Interestingly, the same allele also caused minor retention of RNA Polymerase II (Pol II) and altered nucleosome occupancy across the same region of the gene. These results suggest that specific DNA sequences at the 3’ ends of genes can play roles in promoting efficient yFACT and Pol II dissociation from genes and can also contribute to proper chromatin architecture.
Similar content being viewed by others
Introduction
Whereas the mechanisms that lead to eukaryotic transcription termination and disengagement of RNA Polymerase II (Pol II) from protein-encoding genes are well understood [1], those that control dissociation of transcription elongation factors from DNA following transcription are not as well defined. In previous work using the Saccharomyces cerevisiae model system, we showed that specific amino acid substitutions within the ISGI (Influences Spt16-Gene Interactions) region of the nucleosome cause a shift in occupancy of the transcription elongation factor Spt16 towards the 3’ end of transcribed genes, a defect we have attributed to impaired dissociation of Spt16 following transcription termination [2]. Notably, the strongest ISGI mutants cause only minor perturbations in Pol II-gene occupancy, suggesting that disengagement of Spt16 and Pol II can at least in part be uncoupled from each other [3, 4]. However, other studies have shown that mutant versions of proteins required for proper Pol II dissociation from the 3’ end of genes also partially impair Spt16 dissociation, thus providing evidence that Spt16 dissociation is partially dependent on Pol II termination [5].
Spt16 is a component of the highly conserved histone chaperone complex FACT (FAcilitates Chromatin Transcription/Transactions), which in yeast also contains the protein Pob3 [6,7,8,9]. Results from several groups are consistent with a model in which yeast FACT (yFACT) is first recruited to a gene promoter through interactions with a partially unwrapped + 1 nucleosome and then travels across the gene in a processive manner with assistance from the chromatin remodeler Chd1 [10, 11] to facilitate both reassembly and disassembly of nucleosomes, with the latter activity being more critically important for normal cellular functions [12]. The importance of proper FACT function for organismal health is highlighted by recent work showing a correlation between reduced Spt16 activity and presentation of neurodevelopmental abnormalities in human patients [13]. Similar neurodevelopmental disorders have also been associated with expression of mutant versions of the human histone H3.3 protein – interestingly, one of these mutants (H3.3-L61R), which was identified in separate studies in two different patients [14, 15], corresponds to the strongest ISGI mutant we have identified in yeast [16, 17], thus providing a possible connection between abnormal yFACT dissociation from genes and human disease.
In addition to specific nucleosomal features (e.g., the ISGI region) and other proteins (e.g., proteins that promote Pol II dissociation), it is possible that dissociation of yFACT from genes also requires specific DNA sequences at the 3’ end of genes. For example, DNA sequences could serve as binding sites for proteins that are recruited to termination sequences and subsequently cause yFACT to disengage from the DNA, they could assemble into either particularly stable or unstable nucleosomes (either of which could in turn promote yFACT dissociation), or they could inherently destabilize yFACT-DNA interactions in ways similar to those recently reported for T-tracts in promoting destabilization of Pol II at terminator regions [18]. To test for possible roles for DNA sequences in controlling yFACT dissociation, we used the constitutively expressed gene PMA1 as a model and generated fourteen distinct deletions within its 3’ region and assessed their effects on Spt16 occupancy. We found that one of these deletions caused a modest increase in Spt16 binding, as well as minor retention of Pol II and alterations in nucleosome occupancy, across the 3’ region of the gene. Overall, these results suggest that DNA sequences across termination regions can play roles in promoting dissociation of yFACT and Pol II from genes as well as in maintaining proper chromatin environments.
Results and discussion
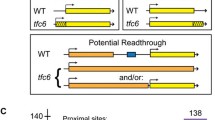
To test for possible contributions of DNA sequences on the dissociation of yFACT from transcribed genes, we generated fourteen distinct 68 base-pair tandem internal deletions across the 3’ region of the PMA1 gene and tested their effects on Spt16 occupancy using the chromatin immunoprecipitation assay followed by quantitative PCR (ChIP/qPCR). PMA1 was chosen for these experiments since it is a highly and constitutively expressed gene and one that we have utilized extensively in past studies when assessing the effects of histone mutants on yFACT-chromatin interactions. The PMA1 internal deletions (diagrammatically shown in Fig. 1A) were constructed through a homologous recombination strategy in strains that had the LEU1 gene – which is located directly downstream from PMA1 and encodes an enzyme required for leucine biosynthesis – either completely or partially replaced with either the URA3 or HIS3 nutritional markers. Transformation of these strains with PCR products harboring regions homologous to the locus but with gaps across their lengths corresponding to the desired deletions (introduced through the design of the forward primers used in the PCR reactions) led to the generation of each internal deletion in situ, reconstitution of the wild-type LEU1 gene (which allowed for selection of the desired recombination events on media lacking leucine), and loss of the URA3 or HIS3 markers. Thus, all the strains assayed in this study (see Table 1) are isogenic to each other except for the presence of distinct 68 base-pairs deletions across the 3’ region of their PMA1 gene (or absence of a deletion in the case of the wild-type control strain).
Assessment of effects of PMA1 3’-internal deletions on Spt16 and Rpb3 occupancy across the 3’ region of PMA1.(A) Cartoon representation of the PMA1-LEU1 region, with arrows indicating the direction of transcription of both genes. Internal deletions within the 3’ region of the PMA1 gene are indicated by the alternating light-blue and purple segments, with their corresponding numbers, 1–14, indicated. The ChIP amplicons represent the regions amplified in the qPCR experiments following the ChIP steps that were used to assess Spt16 and Rpb3 binding. Previously identified PMA1 cleavage/poly-adenylation sites (pA sites, [20]) and the major LEU1 transcription start site (TSS, [23]) are indicated. The approximate region of transcription termination at PMA1 in wild-type cells is also shown: this is based on our previous Pol II ChIP data showing robust Pol II binding at region 3081–3170 and strongly decreased binding at region 3373–3442 [5] which, combined with the location of the most downstream pA site at position 3277, place the approximate region of transcription termination across region 3277–3442. (B) Results from ChIP/qPCR experiments assessing occupancy levels of Spt16 across the PMA1 gene in the wild-type strain (WT) and each of the fourteen strains harboring the indicated internal deletions (ID#). For most strains, regions A, B, and C were assayed, except for the ID10 and ID11 strains, which carry deletions overlapping the B amplicon, for which the BI amplicon was used instead of the B amplicon. In each case, Spt16 occupancy levels are shown relative to its occupancy at the corresponding A region and are presented as mean ± S.E.M from three independent experiments. Statistically significant differences for regions B and C relative to the wild-type strain are indicated by asterisks (Student’s t-test, P < 0.05). For these experiments, chromatin shearing was carried out to achieve enrichment of fragments in the 200–500 base-pair range. (C) Results from ChIP/qPCR experiments assessing binding levels of Rpb3 across PMA1 using the same strains and presented in the same manner as in (B), except that, since the data across repeats tended to be more varied, each result shown represents the mean ± S.E.M from six independent experiments instead of three. The strains used in these experiments are yADP141-155. Detailed information on the primers used in these experiments is provided on Table S1
Following the generation and validation of the strains by Sanger sequencing, we carried out Spt16-ChIP/qPCR assays as previously described [19] to assess Spt16 occupancy at three different locations across the PMA1 gene. As shown in Fig. 1B, thirteen of the fourteen internal deletions did not cause defects in Spt16 occupancy at the 3’ end of PMA1. However, one internal deletion – ID7 – did cause a modest statistically significant increase in Spt16 occupancy over the 3’ region of the gene with respect to its occupancy at the 5’ region (Fig. 1B), suggesting that DNA sequences can play roles in promoting yFACT dissociation from genes. Given our previous work providing evidence that dissociation of Spt16 from transcribed genes is partially dependent on Pol II dissociation [5], we considered the possibility that the Spt16 dissociation defect conferred by ID7 could be a result of abnormal retention of Pol II onto the DNA. To test for this, we assessed occupancy of Rpb3, one of the components of Pol II, across PMA1 in the fourteen internal deletion strains using ChIP/qPCR. Similar to the Spt16 results, only ID7 conferred a statistically significant increase in Rpb3 occupancy at the 3’ end of PMA1 (Fig. 1C). ID7 removes base pairs + 3166 through + 3233 of PMA1 (with + 1 corresponding to the A of the ATG start codon), which are located just upstream from one of two major PMA1 polyadenylation sites (mapped at + 3277 [20]) and likely contain one or both elements normally found upstream from pA sites (i.e., the Efficiency Element (EE) and the Positioning Element (PE)) that are known to facilitate 3’-end formation in yeast [21]) – it is therefore possible that ID7 interferes with Pol II termination, which in turn causes abnormal Spt16 retention at the 3’ end of the gene. We note that the modest increase in retention of Spt16 over the PMA1 3’ region in the context of ID7 is not expected to affect expression of PMA1 since much higher retention levels of Spt16 due to the presence of the ISGI mutants H3-L61W do not cause significant changes in PMA1 expression ([3] and data not shown).
Although ID7 only has a modest effect on Spt16 and Rpb3 dissociation from PMA1, it is possible that abnormally increased occupancy levels of one or both of these factors can cause alterations in chromatin structure across the 3’ end of PMA1. To test this, we used a micrococcal nuclease (MNase)/qPCR strategy [22] to map and compare nucleosome occupancy across this region in wild-type and ID7 strains, as well as in a strain harboring an internal deletion that does not confer defects in Spt16 or Rpb3 binding as an additional control (ID2). In wild-type cells, our assay detects the presence of four nucleosomes across this region, with the two located farther downstream from the PMA1 open reading frame being the most stable ones (Fig. 2). Whereas in the ID2 strain the positioning and stability of all four nucleosomes are indistinguishable from what is seen in the wild-type strain, moderate changes in the chromatin structure across the region are detected in the ID7 strain. More specifically, while ID7 does not affect the two upstream nucleosomes, it does appear to destabilize the two downstream nucleosomes (as suggested by decreased MNase protection), with the one located more upstream potentially also becoming less well-positioned (as suggested by the broader MNase-protection peak) (Fig. 2). Since ID7 removes a region located well upstream from the four nucleosomes detected in our assay, it is unlikely that the deletion is directly responsible for the changes in chromatin structure we observe – rather, the changes in the MNase-sensitivity patterns of the two downstream nucleosomes are likely a consequence of the abnormally higher levels of yFACT and/or Pol II occupancy in the region due to their impaired dissociation caused by ID7. Whereas it is unclear why the two upstream nucleosomes are unaffected by ID7, it is possible that the abnormally retained yFACT complex is positioned just downstream from these nucleosomes, which may consequently be subject to yFACT-mediated reassembly.
Effects of internal deletions 2 (ID2) and 7 (ID7) on nucleosome occupancy across the 3’ region of PMA1. (A) Nucleosome occupancy across the 3’ region of PMA1 in wild-type, ID2, and ID7 strains was assessed using a previously published protocol [22]. Briefly, logarithmically grown cells were spheroplasted and exposed to micrococcal nuclease (MNase), and MNase-resistant DNA derived from single nucleosomes was isolated and amplified using qPCR. The regions quantified by qPCR are indicated in the figure and labeled NP (Nucleosome Positioning) amplicons – the ChIP amplicons used in Fig. 1 are also shown here to facilitate a comparison of the locations assayed in the ChIP and the nucleosome positioning experiments. In each case, protection levels are relative to the protection level measured for a “strong” nucleosome located at the 5’ region of the GAL1 open reading frame. For each of the three strains assayed, the locations of nucleosomes across the PMA1 3’ region shown in the diagrams below the graph are based on the peaks in MNase protection, with darker shades of gray corresponding to higher peaks, or more stable nucleosomes. In each case, the results are presented as mean ± S.E.M from three independent experiments. Since the internal deletions remove 68 base pairs from the PMA1’s 3’ region, the location of the nucleosome in ID2 and ID7 are shifted closer to the end of the PMA1 open reading frame compared to the nucleosomes in wild-type cells, making it difficult to compare MNase protection profiles across the strains. To facilitate comparison across the three strains, the data shown in (A) are reproduced in (B) but the length of the 3’ PMA1 region in the ID2 and ID7 strains is artificially maintained the same as that in wild-type cells by introducing a gap corresponding to 68 base pairs where the corresponding regions have been removed. Representation of the data in this fashion makes it clear that ID7 causes alterations in the occupancy levels of the two more downstream nucleosomes, with statistically significant differences relative to the wild-type strain indicated by asterisks (Student’s t-test, P < 0.05). The strains used in these experiments are yADP141, yADP143, and yADP148. Detailed information on the primers used in these experiments is provided on Table S1
Our results suggest that specific DNA sequences at 3’ ends of genes can play roles in promoting yFACT dissociation from genes. At PMA1 the region encompassed by ID7 appears to play this role, but its mechanism of action remains to be elucidated. Given the observation that this region is also needed for full Pol II dissociation from PMA1 and that Spt16 dissociation is at least in part dependent on Pol II termination [5], it is likely that rather than playing a direct role in controlling yFACT dissociation from chromatin, this region is required for Pol II dissociation and that yFACT’s retention at this region is a consequence of the abnormally higher presence of Pol II on the DNA. Regardless of the mechanism, it is interesting to see that the modest increase in yFACT and Pol II occupancy across the 3’ end of PMA1 causes significant alterations in chromatin structure downstream from where these factors are retained. These changes could be due to the nucleosome disassembly activity carried out by the excess yFACT complex at this location, by subtle run-through transcription by the retained Pol II complex into the LEU1 gene, or the combination of both events. A more general conclusion that can be drawn from these studies is that mutations or conditions that cause even small increases in yFACT and/or Pol II chromatin occupancy can lead to significant changes in the surrounding chromatin environments that can potentially lead to important biological outcomes.
Limitations
Whereas our studies provide evidence that DNA sequences at the 3’ end of PMA1 can play modest roles in promoting yFACT and Pol II dissociation from DNA and can also contribute to normal chromatin structure in the region, additional studies at other genes would need to be carried out to determine if these observations apply more broadly across the genome.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The GenBank accession numbers for the fourteen Internal Deletion sequences are OR451999-OR452012. The reference sequence for the PMA1 3’ region was obtained from the Saccharomyces Genome Database found at https://www.yeastgenome.org.
Abbreviations
- FACT:
-
Facilitates Chromatin Transcription/Transactions
- Pol II:
-
RNA Polymerase II
- Spt16:
-
Suppressor of Ty insertion mutations 16
- ChIP:
-
Chromatin Immunoprecipitation
- MNase:
-
Micrococcal Nuclease
- qPCR:
-
quantitative Polymerase Chain Reaction
References
Rodriguez-Molina JB, West S, Passmore LA. Knowing when to stop: transcription termination on protein-coding genes by eukaryotic RNAPII. Mol Cell. 2023;83(3):404–15.
Nyamugenda E, Cox AB, Pierce JB, Banning RC, Huynh ML, May C, et al. Charged residues on the side of the nucleosome contribute to normal Spt16-gene interactions in budding yeast. Epigenetics. 2018;13(1):1–7.
Duina AA, Rufiange A, Bracey J, Hall J, Nourani A, Winston F. Evidence that the localization of the elongation factor Spt16 across transcribed genes is dependent upon histone H3 integrity in Saccharomyces cerevisiae. Genetics. 2007;177(1):101–12.
Nguyen HT, Wharton W 2nd, Harper JA, Dornhoffer JR, Duina AA. A nucleosomal region important for ensuring proper interactions between the transcription elongation factor Spt16 and transcribed genes in Saccharomyces cerevisiae. G3. (Bethesda). 2013;3(6):929–40.
Campbell JB, Edwards MJ, Ozersky SA, Duina AA. Evidence that dissociation of Spt16 from transcribed genes is partially dependent on RNA polymerase II termination. Transcription. 2019;10(4–5):195–206.
Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92(1):105–16.
Formosa T. FACT and the reorganized nucleosome. Mol Biosyst. 2008;4(11):1085–93.
Zhou K, Liu Y, Luger K. Histone chaperone FACT FAcilitates chromatin transcription: mechanistic and structural insights. Curr Opin Struct Biol. 2020;65:26–32.
Jeronimo C, Robert F. The histone chaperone FACT: a guardian of chromatin structure integrity. Transcription. 2022:1–23.
Jeronimo C, Angel A, Nguyen VQ, Kim JM, Poitras C, Lambert E, et al. FACT is recruited to the + 1 nucleosome of transcribed genes and spreads in a Chd1-dependent manner. Mol Cell. 2021;81(17):3542–e5911.
Martin BJE, Chruscicki AT, Howe LJ. Transcription promotes the Interaction of the FAcilitates chromatin transactions (FACT) complex with nucleosomes in Saccharomyces cerevisiae. Genetics. 2018;210(3):869–81.
Gurova K, Chang HW, Valieva ME, Sandlesh P, Studitsky VM. Structure and function of the histone chaperone FACT - resolving FACTual issues. Biochim Biophys Acta Gene Regul Mech; 2018.
Bina R, Matalon D, Fregeau B, Tarsitano JJ, Aukrust I, Houge G, et al. De novo variants in SUPT16H cause neurodevelopmental disorders associated with corpus callosum abnormalities. J Med Genet. 2020;57(7):461–5.
Bryant L, Li D, Cox SG, Marchione D, Joiner EF, Wilson K et al. Histone H3.3 beyond cancer: germline mutations in histone 3 family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients. Sci Adv. 2020;6(49).
Maver A, Cuturilo G, Ruml SJ, Peterlin B. Clinical next generation sequencing reveals an H3F3A gene as a New Potential Gene Candidate for Microcephaly Associated with severe Developmental Delay, Intellectual disability and growth retardation. Balkan J Med Genet. 2019;22(2):65–8.
Johnson P, Mitchell V, McClure K, Kellems M, Marshall S, Allison MK, et al. A systematic mutational analysis of a histone H3 residue in budding yeast provides insights into chromatin dynamics. G3 (Bethesda). 2015;5(5):741–9.
Pablo-Kaiser A, Tucker MG, Turner GA, Dilday EG, Olmstead AG, Tackett CL, et al. Dominant effects of the histone mutant H3-L61R on Spt16-gene interactions in budding yeast. Epigenetics. 2022;17(13):2347–55.
Han Z, Moore GA, Mitter R, Lopez Martinez D, Wan L, Dirac Svejstrup AB, et al. DNA-directed termination of RNA polymerase II transcription. Mol Cell. 2023;83(18):3253–67. e7.
Myers CN, Berner GB, Holthoff JH, Martinez-Fonts K, Harper JA, Alford S, et al. Mutant versions of the S. Cerevisiae transcription elongation factor Spt16 define regions of Spt16 that functionally interact with histone H3. PLoS ONE. 2011;6(6):e20847.
Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3’ ends of genes. Embo J. 2004;23(2):354–64.
Guo Z, Sherman F. 3’-end-forming signals of yeast mRNA. Trends Biochem Sci. 1996;21(12):477–81.
Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453(7192):246–50.
Hsu YP, Schimmel P. Yeast LEU1. Repression of mRNA levels by leucine and relationship of 5’-noncoding region to that of LEU2. J Biol Chem. 1984;259(6):3714–9.
Acknowledgements
The authors thank Tim Formosa for providing the Spt16-specific antibodies used for the Spt16 ChIP/qPCR and David Addepalli for carrying out pilot experiments that facilitated the execution of the MNase/qPCR experiments reported in this work. We also thank Reine Protacio and current members of the Duina lab for reviewing the manuscript before submission.
Funding
This material is based upon work supported by the National Science Foundation under Grants No. 1613754 and No. 2015806.
Author information
Authors and Affiliations
Contributions
S.E.B., B.H., S.A.O., M.R.N., and L.J. carried out the ChIP/qPCR experiments shown in Fig. 1. A.W.C., D.H., H.P., and C.E.T. participated in the construction of the strains carrying the 14 internal deletions within the 3’ region of the PMA1 gene. A.A.D. carried out the MNase/qPCR experiments shown in Fig. 2, wrote the manuscript, and designed and supervised all aspects of the project. All authors have reviewed the manuscript and approve of its contents.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Byrd, S.E., Hoyt, B., Ozersky, S.A. et al. Assessing contributions of DNA sequences at the 3’ end of a yeast gene on yFACT, RNA polymerase II, and nucleosome occupancy. BMC Res Notes 17, 219 (2024). https://doi.org/10.1186/s13104-024-06872-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-024-06872-y