Abstract
Objective
A Delta-Notch signaling component, Notch1, is involved in the normal development and multiple disorders of the kidney. Although the increase in Notch1 signaling is crucial to these pathogeneses, the basal signaling level in ‘healthy’ mature kidneys is still unclear. To address this question, we used an artificial Notch1 receptor fused with Gal4/UAS components in addition to the Cre/loxP system and fluorescent proteins in mice. This transgenic reporter mouse system enabled labeling of past and ongoing Notch1 signaling with tdsRed or Cre recombinase, respectively.
Results
We confirmed that our transgenic reporter mouse system mimicked the previously reported Notch1 signaling pattern. Using this successful system, we infrequently observed cells with ongoing Notch1 signaling only in Bowman’s capsule and tubules. We consider that Notch1 activation in several lines of disease model mice was pathologically significant itself.
Similar content being viewed by others
Introduction
The Delta-Notch signaling pathway is a ligand‒receptor signaling pathway that mediates cell‒cell communication in developing organs [1]. Upon binding with Delta ligands, the Notch receptor intracellular domain is cleaved and acts as a transcription factor [2]. This signaling pathway is involved in multiple disorders as well as developmental processes. For example, Notch1 is reportedly involved in acute T-cell lymphoblastic leukemia [3], non-small cell lung carcinomas [4], and diabetic foot ulcerations [5]. In the kidney, Notch1 activation was observed in podocytes and tubulointerstitial cells from human patients with diabetic kidney disease (DKD) and focal segmental glomerular sclerosis (FSGS) [6]. Although the involvement of Notch1 in these disorders has been validated, its cellular mechanism is controversial. On the one hand, the Notch1 intracellular domain was expressed in podocytes of DKD and FSGS patients [7]. On the other hand, Notch1 was expressed in endothelial cells in the glomeruli, and overexpression of the Notch1 intracellular domain in Tie2-expressing endothelial cells led to albuminuria via decreased VE-cadherin expression [8]. To further characterize the involvement of Notch1 activity in these disorders, it is necessary to know the basal Notch1 activity in healthy subjects. A previous study suggested that Notch1 activity in the ‘healthy’ mature kidney was very low using a transgenic mouse line (N1IP::Cre(LO), formerly known as NIP-CRE) that expressed Notch1-Cre fusion protein [9]. The same group, however, generated the second‒generation Notch1-Cre mouse (N1IP::CreHI), which improved the detection sensitivity of Notch1 activity to demonstrate past Notch1 signaling in epithelial cells from the proximal to distal tubule of the mature kidney [10]. Indeed, single-cell RNA-seq revealed that Notch1 signaling promoted the maturation of all nephron segments and selection of proximal tubular cell fate [11]. In contrast to its developmental contribution, ongoing Notch1 signaling in the ‘healthy’ mature kidney is still unclear.
Here, we examined ongoing Notch1 signaling in mature kidneys using our original reporter mouse system that consisted of three transgenic mouse lines. The first mouse line expresses the artificial Notch1 receptor, the intracellular domain of which was replaced with the yeast transcription factor Gal4VP16 (N1-Gal4VP16 mouse) [12]. The second transgenic mouse line expresses Cre recombinase when Gal4VP16 binds to its upstream activating sequence (UAS) (UAS-Cre mouse) [13]. These two mouse lines enable visualization of ongoing Notch1 signaling by immunohistochemistry for Cre recombinase. The third mouse line expresses EGFP or tandem dsRed (tdsRed) before and after Cre-mediated recombination, respectively. In detail, the tdsRed coding sequence was inserted into the ROSA26 locus following a floxed EGFP-STOP sequence (R26GRR mouse) [14]. Therefore, past and ongoing Notch1 signals are labeled with tdsRed or Cre recombinase, respectively (Fig. 1).
TdsRed protein was detected in proximal tubular epithelial cells, suggesting that our reporter mouse system worked. Using this working system, we examined ongoing Notch1 signaling in the ‘healthy’ mature kidney.
Materials and methods
Animal Welfare
Animal experiments were carried out in accordance with the Regulation for Animal Experiments in our university and Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology. Approval was obtained from the Institutional Animal Care and Use Committee and the DNA Experiment Committee of the University of Tsukuba (Approval Numbers for Animal Experiments: 22–059) (Approval Number for DNA Experiments: 220018). UAS-Cre mice are available from RIKEN through the National BioResource Project of Japan: Crl:CD1(ICR)-Tg(UAS-cre/T2A/miRFP670)216Staka (No. RBRC11716). In addition, R26GRR mice are also available from RIKEN through the National BioResource Project of Japan: C57BL/6 N-Gt(ROSA)26Sor < tm1(CAG-EGFP/tDsRed)Utr>/Rbrc (No. RBRC04874).
Histological analysis
We used three N1-Gal4VP16; UAS-Cre; R26GRR mice (10–13 weeks old), one UAS-Cre; R26GRR mouse (6 weeks old) and one Non-Tg mouse (5 weeks old) for the analysis. To reduce the number of mice used, we used post-weaning single mouse per group for the negative control (UAS-Cre; R26GRR and Non-Tg groups). Prior to sampling the organs, mice were sacrificed by cervical dislocation and perfused with PBS and Mildform 10 N (Cat #: 133-10311, Fujifilm, Osaka, Japan). For fluorescent imaging, 10-µm-thick frozen sections were counterstained with Hoechst 33342 (Cat #: H3570, Invitrogen, Waltham, MA, USA). For immunohistochemistry, 4-µm-thick paraffin sections were sequentially incubated with a rabbit monoclonal anti-Cre recombinase antibody (clone: D7L7L, Cat #: 15036 S, RRID: AB_2798694, Cell Signaling Technology) or a rabbit anti-RFP antibody (cat #: 600-401-379, Lot #: 46317, RRID: AB_2209751, Rockland Immunochemicals, Philadelphia, PA, USA), Histofine SimpleStain (Cat #: 414341, Nichirei Biosciences, Tokyo, Japan), Histofine DAB Substrate Kit (Cat #: 425011, Nichirei Biosciences) and counterstained with Mayer’s hematoxylin solution (Cat #: 131–09665, Fujifilm, Osaka, Japan). Images were captured by using BIOREVO-BZ-X810 (Keyence, Osaka, Japan).
Results and discussion
Examination of past notch1 signaling in ‘healthy’ mature kidneys
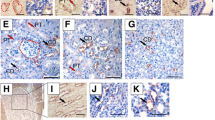
To examine whether our reporter mouse system works, we started by visualizing the past Notch1 signal (tdsRed expression) in the ‘healthy’ mature kidney. Fluorescent imaging showed tdsRed expression in the cortex and medulla specifically in N1-Gal4VP16; UAS-Cre; R26GRR mice (Fig. 2a). We observed tdsRed fluorescence in the tubular epithelium and in the glomeruli (Fig. 2b). Furthermore, immunohistochemistry for dsRed protein showed that most of the proximal tubular epithelial cells and podocytes were positive, in concordance with previous reports [10, 11], suggesting that our reporter mouse system worked (Fig. 2c).
Visualization of past Notch1 signaling in ‘healthy’ mature kidneys
(a) Fluorescent examination of ‘healthy’ mature kidney at a low magnification. Note that tdsRed was expressed in both the cortex and medulla specifically in N1-Gal4VP16; UAS-Cre; R26GRR mice. (b) Fluorescent examination of ‘healthy’ mature kidney at a high magnification. Note that tdsRed was expressed in the tubules. (c) Immunohistochemistry for dsRed showed that proximal tubular epithelial cells (arrowheads) and podocytes (arrow) had a Notch1 signal in concordance with previous reports.
Examination of ongoing notch1 signaling in ‘healthy’ mature kidneys
Next, we examined ongoing Notch1 signaling using our reporter mouse system. Overall, Cre-expressing cells were very rare (less than 1% of total kidney cells). Indeed, we never observed Cre-expressing cells in the medulla. In contrast, we occasionally observed these cells in the tubular epithelium (Fig. 3a) and Bowman’s capsule (Fig. 3b). As such, Bowman’s capsule has a pillar or cuboidal epithelium, and Notch1 signaling might be induced following activation of Bowman’s capsule. Collectively, we consider that Notch1 activation in several lines of disease model mice [6,7,8] was pathologically significant itself.
Finally, to examine our finding that Notch1 signaling in ‘healthy’ mature kidney is rare, we conducted a single-cell RNA-sequencing analysis using a previously reported dataset (GSE107585) [15]. The methods and results were described in “Additional file 1”, and the source code used to generate the results is available at GitHub repository (https://github.com/MasaharuYoshihara/Notch1-Signal-Kidney).
Conclusion
We confirmed that proximal tubular epithelial cells underwent Notch1 signaling. In contrast, cells with ongoing Notch1 signaling were very rare in ‘healthy’ mature kidneys.
Limitations
We were unable to describe the specific characteristics of Cre-expressing cells, although those cells tended to be located near each other. That was one reason the present study lacks explanation on the mechanism behind ongoing Notch1 signaling in the ‘healthy’ mature kidney.
Data Availability
The datasets generated and/or analyzed during the current study are available in the figshare repository, https://doi.org/10.6084/m9.figshare.21331212.v1.
Abbreviations
- DKD:
-
diabetic kidney disease
- FSGS:
-
focal segmental glomerular sclerosis
- tdsRed:
-
tandem dsRed
- UAS:
-
upstream activating sequence
References
Collier JR, Monk NAM, Maini PK, Lewis JH. Pattern formation by lateral inhibition with feedback: a mathematical model of Delta-Notch intercellular signalling. J Theor Biol. 1996;183(4):429–46.
Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93(4):649– 60.
Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–61.
George J, Lim JS, Jang SJ, Cun Y, Ozretia L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53.
Zheng X, Narayanan S, Sunkari VG, Eliasson S, Botusan IR, Grünler J, et al. Triggering of a Dll4-Notch1 loop impairs wound healing in diabetes. Proc Natl Acad Sci U S A. 2019;116(14):6985–6994.
Murea M, Park JK, Sharma S, Kato H, Gruenwald A, Niranjan T, et al. Expression of notch pathway proteins correlates with albuminuria, glomerulosclerosis, and renal function. Kidney Int. 2010;78(5):514–22.
Niranjan T, Bielesz B, Gruenwald A, Ponda MP, Kopp JB, Thomas DB, et al. The notch pathway in podocytes plays a role in the development of glomerular disease. Nat Med. 2008;14(3):290–8.
Li L, Liu Q, Shang T, Song W, Xu D, Allen TD et al. Aberrant activation of Notch1 signaling in glomerular endothelium induces albuminuria. Circ Res. 2021;602–18.
Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J et al. Mapping the consequence of Notch 1 proteolysis in vivo with NIP-CRE. Development. 2007;134(3):535–44.
Liu Z, Brunskill E, Boyle S, Chen S, Turkoz M, Guo Y et al. Second-generation Notch1 activity-trap mouse line (N1IP::CreHI) provides a more comprehensive map of cells experiencing Notch1 activity. Development. 2015;142(6):1193–202.
Duvall K, Crist L, Perl AJ, Pode Shakked N, Chaturvedi P, Kopan R. Revisiting the role of Notch in nephron segmentation confirms a role for proximal fate selection during mouse and human nephrogenesis. Development. 2022;149(10).
Smith E, Claudinot S, Lehal R, Pellegrinet L, Barrandon Y, Radtke F. Generation and characterization of a Notch1 signaling-specific reporter mouse line. Genesis. 2012;50(9):700–10.
Yoshihara M, Nishino T, Sambe N, Nayakama T, Radtke F, Mizuno S, et al. Generation of a Gal4-dependent gene recombination and illuminating mouse. Exp Anim. 2022;71(3):385–90.
Hasegawa Y, Daitoku Y, Sekiguchi K, Tanimoto Y, Mizuno-Iijima S, Mizuno S, et al. Novel ROSA26 cre-reporter knock-in C57BL/6 N mice exhibiting Green Emission before and Red Emission after cre-mediated recombination. Exp Anim. 2013;62(4):295–304.
Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–63.
Acknowledgements
The authors are grateful to all members of the Department of Anatomy and Embryology, Institute of Medicine, University of Tsukuba, and American Journal Experts (https://www.aje.com/) for English language editing.
Funding
This work was supported by the PhD Program in Humanics, University of Tsukuba (Doctoral Program for World-leading Innovative and Smart Education, Ministry of Education, Culture, Sports, Science and Technology, Japan), by the Japan Society for the Promotion of Science through JSPS KAKENHI Grant-in-Aid for Research Activity Start-up (Grant number: JP22K20734), and by YOKOYAMA Foundation for Clinical Pharmacology (Grant number: YRY-2207). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of these funding agencies.
Author information
Authors and Affiliations
Contributions
RS made substantial contributions to the conception, performed single-cell RNA-sequencing analysis, and drafted the work. TN1 performed single-cell RNA-sequencing analysis and histological analysis. TN2 helped single-cell RNA-sequencing analysis and made substantial contributions to the histological analysis. NS made substantial contributions to the histological analysis. FR helped the design of the work and substantially revised the draft. MY made substantial contributions to the design of the work, data acquisition (histological analysis and single-cell RNA-sequencing analysis), and interpretation of the data. ST substantially revised the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Animal experiments were carried out in accordance with the Regulation for Animal Experiments in our university and Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology. Approval was obtained from the Institutional Animal Care and Use Committee and the DNA Experiment Committee of the University of Tsukuba (Approval Numbers for Animal Experiments: 22-059) (Approval Number for DNA Experiments: 220018).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sugiura, R., Nakayama, T., Nishino, T. et al. Notch1 signaling is limited in healthy mature kidneys in vivo. BMC Res Notes 16, 54 (2023). https://doi.org/10.1186/s13104-023-06326-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-023-06326-x