Abstract
Background
HIV care programs in resource-limited settings have hitherto concentrated on antiretroviral therapy (ART) access, but HIV drug resistance is emerging. In a cross-sectional study of HIV-positive adults on ART for ≥6 months enrolled into a prospective cohort in Uganda, plasma HIV RNA was measured and genotyped if ≥1000 copies/ml. Identified Drug resistance mutations (DRMs) were interpreted using the Stanford database, 2009 WHO list of DRMs and the IAS 2014 update on DRMs, and examined and tabulated by ART drug classes.
Findings
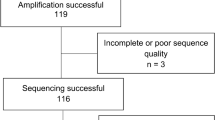
Between July 2013 and August 2014, 953 individuals were enrolled, 119 (12.5%) had HIV-RNA ≥1000 copies/ml and 110 were successfully genotyped; 74 (67.3%) were on first-line and 36 (32.7%) on second-line ART regimens. The predominant HIV-1 subtypes were D (34.5%), A (33.6%) and Recombinant forms (21.8%). The commonest clinically significant major resistance mutations associated with the highest levels of reduced susceptibility or virological response to the relevant Nucleoside Reverse Transcriptase Inhibitor (NRTI) were; the Non-thymidine analogue mutations (Non-TAMS) M184V—20.7% and K65R—8.0%; and the TAMs M41L and K70R (both 8.0%). The major Non-NRTI (NNRTI) mutations were K103N—19.0%, G190A—7.0% and Y181C—6.0%. A relatively nonpolymorphic accessory mutation A98G—12.0% was also common. Seven of the 36 patients on second line ART had major Protease Inhibitor (PI) associated DRMS including; V82A—7.0%, I54V, M46I and L33I (all 5.0%). Also common were the accessory PI mutations L10I—27%, L10V—12.0% and L10F—5.0% that either reduce PI susceptibility or increase the replication of viruses containing PI-resistance mutations. Of the 7 patients with major PI DRMs, five had high level resistance to ritonavir boosted Lopinavir and Atazanavir, with Darunavir as the only susceptible PI tested.
Conclusions
In resource-limited settings, HIV care programs that have previously concentrated on ART access, should now consider availing access to routine HIV viral load monitoring, targeted HIV drug resistance testing and availability of third-line ART regimens.
Similar content being viewed by others
Background
Antiretroviral therapy (ART) improves survival and quality of life of HIV-infected individuals and controls HIV transmission, however HIV-1 drug resistance limits the benefits of ART [1].
Globally, the number of HIV-positive people is increasing and the numbers of new HIV infections and mortality due to AIDS has declined. In 2014, globally 14.9 million (40%) people living with HIV were receiving ART, of which 13.5 million were in low- and middle-income countries [2]. In Uganda, 750,896 (50%) HIV infected people were receiving ART by December 2014 [3].
Despite the progress in the rapid ART scale up, HIV drug resistance profiling prior to starting ART, and routine virological monitoring and drug resistance testing are not yet standard HIV care in resource limited settings as is the case in the developed world [4]. The emergence of HIV drug resistance may limit the sustained benefits of ART in settings with limited laboratory monitoring and drug options. Drug resistance prevalence varies widely, at 2.8% in sub-Saharan Africa compared to 11.5% in North America, while in South Africa rising levels of acquired antiretroviral drug resistance and newly infected patients with resistant viruses have been reported [5, 6].
Although HIV care programs in resource limited settings have hitherto concentrated on expanding ART access, the emergency of HIV drug resistance is a challenge. We document the antiretroviral (ARV) drug resistance mutations and susceptibility patterns among Ugandan adults on ART. We also highlight the need for HIV care programs in resource limited settings to avail access to routine virological monitoring, access to targeted HIV drug resistance testing and alternative third line ART regimen drugs.
This was a cross-sectional study of HIV-positive adults (18 years and above) at enrolment into a prospective clinical cohort established to study the complications of long-term ART (CoLTART). Individuals on ART for 6 months or more were recruited from two ART cohorts; the Entebbe site of the former Development of Antiretroviral Therapy in Africa (DART) Trial cohort established in 2003 [7], and the former Rural Clinical Cohort in south western Uganda where ART was introduced in 2004 [8]. ART history was obtained from the former ART cohorts and included; dates of ART initiation and switches, ART regimens, baseline viral load and CD4 cell counts. First-line ART regimens consisted of two nucleoside reverse transcriptase inhibitors (NRTI) and one non-nucleoside reverse transcriptase inhibitor (NNRTI). Some participants in the DART Trial initiated ART with a triple nucleoside (3 NRTIs) first-line ART regimen. Second-line ART regimens consisted of one or two NRTIs and a boosted protease inhibitor (PI). Individual ARVs in an ART regimen were substituted in the event of adverse drug effects, contraindications by medical conditions or interactions with other concurrently administered medications while complete ART switches were made in the event of treatment failure.
CD4 cell counts were measured with the FACSCount or FACSCalibur machine (Becton–Dickinson, USA). Plasma HIV-1 RNA was quantified using the Cobas Ampliprep/Cobas Taqman 48 version 2.0 HIV-1 viral load assay, [Roche Molecular Diagnostics (RMD), NJ, USA]. Samples with HIV RNA viral load of 1000 copies/mL or higher were genotyped. For ARV drug resistance testing, HIV RNA was extracted using the QIAmp Viral RNA mini kit (Qiagen), the entire protease and amino terminus of the reverse transcriptase was amplified, cleaned and sequenced using the ABI 3500 machine (Applied Biosystems). Sequences were base-called using Sequencher v5.2.4 and sequence alignments performed using BioEdit v7.2.5 [9] and SeaView v4.0 [10]. HIV-1 subtyping was done using SCUEAL and REGA online software [11], and Recombinant Identification Program (RIP). Drug resistance mutations (DRMs) were interpreted using the Stanford HIVdb Program. Assigned DRMs were interpreted using the 2009 WHO list for epidemiological surveillance of TDR alongside the lAS 2014 Update of DRMs of HIV-1. Basic phylogenies were performed to determine sequence relatedness and to rule out contaminations. Sequences with genetic mixtures of wild-type and mutant sequences at amino acid sites that code for SDRMs were considered to be drug-resistant. Quality Assurance was done using the Calibrated Population Resistance tool (CPR), Stanford and the Los Alamos National database (LANL dbase) for the HIV Sequence Quality Analysis [12]. The Laboratory is WHO HIVDR Region accredited.
Participants with HIV-1 RNA viral load of 1000 copies/mL or higher, whose samples were successfully amplified and genotyped were included in this analysis. The socio-demographic characteristics including age and sex were examined and tabulated. ART data consisted of the type of ART regimen and duration on ART. The three ART regimen types were; Triple nucleoside (3 NRTIs), 2 NRTIs with a NNRTI, and a PI-based regimen. Duration on ART was grouped into below 5 years, 5–9 years, and above 9 years. Drug resistance mutations to one antiretroviral class only (either NRTI, NNRTI or PI), two classes only (either NRTI and NNRTI, PI and NRTI, or PI and NNRTI) or three antiretroviral classes (NRTI and 1 NNRTI and PI) were tabulated. For participants with a major PI DRM, we showed the resistance levels to each antiretroviral drug.
Findings
Out of the 953 individuals on ART for 6 or more months, 119 (12.5%) had a viral load ≥1000 copies/ml. The five samples that did not amplify and the 4 with insufficient quantities were excluded from this analysis. Of the 110 individuals whose samples were sequenced for drug resistance testing, 73 (66.4%) were females, 95 (86.4%) were aged 35 years and above, 89 (80.9%) had been on ART for 9 or more years, with 74 (67.3%) on first and 36 (32.7%) on second-line ART regimens at enrolment. Sixty-five (59.1%) had CD4 cell counts of 350 cells/ml or less and 67 (60.9%) had HIV RNA of 10,000 copies/ml or more. The predominant HIV-1 subtypes were D (34.5%), A (33.6%) and Recombinant forms (21.8%) (Table 1). Of the 110 individuals with drug resistance testing results, 8 (7.3%) had no detectable drug resistance mutations (DRMs) while 102 (92.7%) had at least one detectable DRMs, distributed as follows: DRMs to any NRTI—92 (83.6%), to any NNRTIs—77 (70.0%) and to any PIs—33 (30.0%). DRMs by ART regimen line were: triple nucleoside—39 (35.5%), 2NRTI with an NNRTI—31 (28.2%) and second-line PI based regimen—33 (30.0%). Twenty-two participants (20.0%) had DRMs to one ARV class only (NRTI—14, NNRTI—7 and PI—1), 60 (54.5%) participants had dual-class DRMs (NRTI + NNRTI—48, PI + NRTI—10 and PI + NNRTI—2, while 20 (18.2%) had triple class DRMs (Table 2).
In our cohort, the commonest clinically significant major NRTI resistance mutations associated with highest levels of reduced susceptibility or virological response were the Non-thymidine analogue mutations (Non-TAMs): M184V—20.7% and K65R—8.0%, while M41L and K70R (both 8.0%) were the commonest TAMs. The commonest major NNRTI resistance mutations known to reduce susceptibility or virological response to NNRTIs were: K103N—19.0%, G190A—7.0% and Y181C—6.0%. A relatively nonpolymorphic accessory mutation A98G—12.0% was also common. Of the 33 identified PI resistance mutations, 7 were major mutations and 26 minor mutations. The most common major PI-resistance mutations associated with the highest levels of phenotypic resistance were: V82A—7.0% and I54V, M46I, L33I (all 5.0%). The identified accessory PI resistance mutations that either reduce PI susceptibility or increase the replication of viruses containing PI-resistance mutations included: L10I—27.0%, L10V—12.0% and L10F—5.0% (Table 2).
Of the 7 patients with major PI DRMs, high level resistance to Indinavir (IDV), Fosamprenavir (FPV), Lopinavir (LPV) and Nelfinavir (NFV)—to each was found among 4 patients. Intermediate level drug resistance to Saquinavir (SQV) was found among 4 patients, Tipranavir (TPV) and IDV among 3 patients. The HIV among six of the 7 patients with major PI mutations were susceptible to Darunavir with one expressing low level resistance to this drug, and two expressing susceptibility to Tipranavir (Table 3).
Conclusions
We found that nearly half of our patients with virological failure had resistance to both NRTIs and NNRTIs, about a fifth had resistance to the three classes of ARVs commonly used in Uganda while a small proportion had no Drug Resistance Mutations (DRMs). NRTIs have fewer side effects and toxicity than NNRTIs, but drug resistance could diminish their efficacy. As expected in Africa, the predominant NRTI mutation observed was M184V [13]. Type 1 TAM M41L causes higher levels of phenotypic and clinical resistance to thymidine analogues and cross resistance to Abacavir, Tenofovir and Didanosine than Type 11 K70R. The M184V and K65R DRMs might be due to the Tenofovir and Lamivudine ART backbone and the triple nucleoside regimen of Abacavir-Zidovudine-Lamivudine that most of our patients were initiated on. Triple nucleoside first-line ART regimens have since been discontinued as they are virologically inferior to a regimen containing Efavirenz (NNRTI) plus two or three NRTIs [14]. The NNRTI mutations observed included K103N, G190A and Y181C which cause high level resistance to Nevirapine and variable resistance to Efavirenz [15].
The observed PI resistance mutations (V82A, V82F, V82S, M46I, M46L and I54V) are clinically significant because they are associated with highest levels of phenotypic resistance and/or strongest evidence for interfering with successful PI therapy. Among the seven patients, these mutations conferred high level resistance to ritonavir boosted Lopinavir and Atazanavir, which are the readily available PIs for second-line ART regimens in our setting. Darunavir, the only susceptible PI tested is still expensive and unavailable in public ART centers in Uganda where PIs are reserved for second-line ART regimens. The emergence of HIV drug resistance is inevitable, owing to the high replication and mutation rates of HIV and ART being a life-long treatment. Therefore, as more people are switched to second-line ART, cases of second-line drug failure will increase and necessitate access to third-line or salvage regimens. In Uganda, routine viral load monitoring is not yet standard care, making early identification of treatment failures to prevent transmission of DRMs impossible. Uganda is yet to recommend HIV genotyping prior to ART initiation, therefore diagnosing transmitted HIV drug resistance is impossible. The strength of our study is the long duration on ART among our participants. A source of selection bias might be that some patients with virological failure might have died before enrolment. We also had limited information on confounders like adherence so our findings may not be generalizable. In conclusion, HIV drug resistance is a major challenge for HIV care programs in resource limited settings that have hitherto concentrated on increasing ART access. Therefore, ART programs in these settings should avail routine HIV viral load monitoring for prompt detection of virological failures, targeted HIV drug resistance testing to detect HIV drug resistance among ART failure as well as access to third-line ARV drugs like Darunavir.
Abbreviations
- HIV:
-
human immunodeficiency virus
- ART:
-
antiretroviral therapy
- RNA:
-
ribonucleic acid
- DRM:
-
drug resistance mutation
- WHO:
-
World Health Organisation
- IAS:
-
International AIDS Society
- NRTI:
-
nucleoside reverse transcriptase inhibitors
- NNRTI:
-
non nucleoside reverse transcriptase inhibitors
- PI:
-
protease inhibitors
- AIDS:
-
acquired immune deficiency syndrome
- ARV:
-
antiretroviral
- CoLTART:
-
Complications of Long-Term Antiretroviral Therapy
- DART:
-
development of antiretroviral therapy in Africa
- RIP:
-
recombinant identification program
- TDR:
-
transmitted drug resistance
- IAS:
-
International AIDS Society
- CPR:
-
calibrated population resistance tool
References
Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther. 2013;18(1):115.
WHO, HIV/AIDS. Media centre. 2015. http://www.who.int/mediacentre/factsheets/fs360/en/. Accessed 24 Sept 2015.
UNAIDS, The HIV and AIDS Uganda Country Progress Report. 2014. http://www.unaids.org/sites/default/files/country/documents/UGA_narrative_report_2015.pdf. Accessed 6 Oct 2015.
Kumarasamy N, Krishnan S. Beyond first-line HIV treatment regimens: the current state of antiretroviral regimens, viral load monitoring, and resistance testing in resource-limited settings. Curr Opin HIV AIDS. 2013;8(6):586–90.
Barth RE, et al. Virological follow-up of adult patients in antiretroviral treatment programmes in sub-Saharan Africa: a systematic review. Lancet Infect Dis. 2010;10(3):155–66.
Ali A, et al. Molecular basis for drug resistance in HIV-1 protease. Viruses. 2010;2(11):2509–35.
Mugyenyi P, et al. Routine versus clinically driven laboratory monitoring of HIV antiretroviral therapy in Africa (DART): a randomised non-inferiority trial. Lancet. 2010;375(9709):123–31.
Morgan D, et al. An HIV-1 natural history cohort and survival times in rural Uganda. AIDS. 1997;11(5):633–40.
Hall T. BioEdit: Biological sequence alignment Editor for Windows 95/98/NT/2000/XP/7; Copyright © 1997–2013 Tom Hall, Ibis Biosciences, Carlsbad, CA 92008. http://www.mbio.ncsu.edu/BioEdit/bioedit.html. Accessed 12 Sept 2015.
Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–4.
Bioafrica and R. Institute, HIV Bioinformatics. REGA HIV-1 & 2 Automated Subtyping Tool (Version 2.0). http://www.bioafrica.net/rega-genotype/html/subtypinghiv.html. Accessed 14 Sept 2015.
LANL, Los Alamos National Laboratory. HIV sequence database. Recombinant Identification Program (RIP). http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html. Accessed 20 May 2015.
Ssemwanga D, et al. Update on HIV-1 acquired and transmitted drug resistance in Africa. AIDS reviews. 2014;17(1):3–20.
Gulick RM, et al. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med. 2004;350(18):1850–61.
Tambuyzer L, et al. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir Ther. 2009;14(1):103–9.
Authors’ contributions
BNM, PM, PoK conceived and designed the study; IN, BNM, PaK, JL,FL, AAK participated in data collection; FL, AAK, PoK conducted the laboratory analyses; IN, BNM, JL, drafted the initial manuscript; IK compiled and analysed data; IN, IK, BNM, JL, FL, AAK, PoK, PM interpreted the analysis; all authors contributed to manuscript revisions. All authors read and approved the final manuscript.
Acknowledgements
We acknowledge the contribution of CoLTART study participants, the CoLTART study team, and staff of the Clinical Diagnostic Laboratory Services, Basic Science and Statistics section.
The CoLTART study team Kyamulibwa study site Billy N. Mayanja, Judith Nalwadda, Gladys Nakibuuka, Harriet Namugenyi, Patrick Kazooba, Rosemary Lubega. Entebbe study site Annet Mugisha, Apophia Tereka, Apuuli Kalyebara, Arthur Namara, Diana Nakitto, Deus Wangi, Fred Nume, George Ssemwanga, Gertrude Nabulime, Gladys Nassuna, Gloria Lubega, Ivan Namakoola, Joseph Lutaakome, Lillian Generous, Lydia Matama, Rosemary Massa, Salome Tino, William Nakahima. Basic Science Virology Anne A. Kapaata, Brian Magambo, Chris Parry, Frederick Lyagoba, Jamirah Nazziwa, Maria Nannyonjo. Clinical Diagnostic Laboratory Services Edward Muhigirwa, Faith Wamalugu, Florence Kabajuma, Hope Grania Nakazibwe, Jackson Were, Joan Bwandinga, Juliet Bukenya, Member Zephyrian Kamushaaga, Peter Hughes, Peter Nkurunziza, Priscilla Agatha Balungi, Simon Mukasa, Sureyah Nassimbwa, Tobias Vudriko, William Senyonga, Willyfred Ochola. Statistics Annet Nakimbugwe, Catherine Nampewo, Doreen Nambuba, Erima Naphtali, Grace Barigye, Irene Nakamanya, Ivan Kasamba, Jonathan Levin, Joseph Kahwa, Joy Namutebi Matovu, Lillian Namayirira, Ruth Namulindwa Lubega, Sandra Nabalayo, Solomon Kaddu. Principal Investigator Paula Munderi.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data will not be shared publicly due to the data sharing policy of the MRC/UVRI Uganda Research Unit on AIDS, which requires a prior data sharing agreement. However, a full Data set on the CoLTART study containing the data supporting the study findings in this report can be obtained from the Director, by email to: mrc@mrcuganda.org or the corresponding author.
Ethics approval and consent to participate
The study was approved by the Research and Ethics Committee of the Uganda Virus Research Institute, and the Uganda National Council for Science and Technology. Participants gave informed signed (or witnessed thumb-printed) written consent to participate in the study and confidentiality was ensured throughout the study.
Funding
This research was jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordant agreement.
Author information
Authors and Affiliations
Consortia
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Namakoola, I., Kasamba, I., Mayanja, B.N. et al. From antiretroviral therapy access to provision of third line regimens: evidence of HIV Drug resistance mutations to first and second line regimens among Ugandan adults. BMC Res Notes 9, 515 (2016). https://doi.org/10.1186/s13104-016-2309-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-016-2309-7