Abstract
Background
The autonomous retroelement Long Interspersed Element-1 (LINE-1) mobilizes though a copy and paste mechanism using an RNA intermediate (retrotransposition). Throughout human evolution, around 500,000 LINE-1 sequences have accumulated in the genome. Most of these sequences belong to ancestral LINE-1 subfamilies, including L1PA2-L1PA7, and can no longer mobilize. Only a small fraction of LINE-1 sequences, approximately 80 to 100 copies belonging to the L1Hs subfamily, are complete and still capable of retrotransposition. While silenced in most cells, many questions remain regarding LINE-1 dysregulation in cancer cells.
Results
Here, we optimized CRISPR Cas9 gRNAs to specifically target the regulatory sequence of the L1Hs 5’UTR promoter. We identified three gRNAs that were more specific to L1Hs, with limited binding to older LINE-1 sequences (L1PA2-L1PA7). We also adapted the C-BERST method (dCas9-APEX2 Biotinylation at genomic Elements by Restricted Spatial Tagging) to identify LINE-1 transcriptional regulators in cancer cells. Our LINE-1 C-BERST screen revealed both known and novel LINE-1 transcriptional regulators, including CTCF, YY1 and DUSP1.
Conclusion
Our optimization and evaluation of gRNA specificity and application of the C-BERST method creates a tool for studying the regulatory mechanisms of LINE-1 in cancer. Further, we identified the dual specificity protein phosphatase, DUSP1, as a novel regulator of LINE-1 transcription.
Similar content being viewed by others
Introduction
Long Interspersed Element-1 (LINE-1) is the only autonomous mobile element in the human genome. Over millions of years, LINE-1 sequences have accumulated in our DNA through the process of retrotransposition, entailing the copy and paste of an RNA intermediate [1, 2]. An estimated 500,000 copies of LINE-1 exist in the human genome, complicating the study of LINE-1 and its retrotransposition [3]. Yet, the majority of LINE-1 sequences are non-functional due to 5’ truncations, mutations, and inversions. These nonfunctional sequences are predominantly ancestral LINE-1 sequences, including subfamilies L1PA2-L1PA7, which can no longer mobilize [1, 4, 5]. The remaining 80–100 full length LINE-1 sequences belong to the human specific LINE-1 subfamily (L1Hs) and are capable of retrotransposition, carrying the potential to reshape our genome, alter gene expression, and disrupt genome integrity [6,7,8].
LINE-1 consists of a 5’ UTR, two open reading frames encoding ORF1p and ORF2p proteins, and a 3’UTR with a polyA tail [9, 10]. A fully downstream sense promoter is located within the 5’UTR, controlling LINE-1 mRNA expression [11]. Additionally, an antisense promoter has been identified within the 5’UTR that has been shown to control expression of a third open reading frame on the antisense strand, ORF0, as well as alternative antisense transcript expression [12,13,14]. Throughout the evolution of LINE-1 sequences, the 5’UTR has acquired new regulatory sequences, exhibiting a rapid evolution of host factor binding, especially KRAB zinc finger binding proteins [15]. LINE-1 encoded proteins, ORF1p and ORF2, have remained relatively conserved and both are instrumental in retrotransposition [16, 17]. ORF1p is a nucleic acid chaperone that forms homotrimers and binds LINE-1 mRNA [18, 19]. ORF1p has also been shown to bind ssDNA as well as non-LINE-1 mRNAs [20, 21]. ORF2p’s endonuclease and reverse transcriptase domains provide the enzymatic activity necessary for retrotransposition [22, 23]. Once expressed, ORF1p and ORF2p bind LINE-1 mRNA forming the ribonucleoprotein (RNP). Upon nuclear breakdown during mitosis, the RNP enters the nucleus where the ORF2p endonuclease nicks the DNA and creates a new copy of LINE-1 through target-primed reverse transcription (TPRT) [24,25,26,27]. LINE-1 has also been shown to retrotranspose in non-dividing cells, suggesting an additional mode of entry into the nucleus [28].
Many mechanisms have evolved to silence LINE-1 expression in somatic cells, limiting its potential to mobilize. DNA methylation, histone modifications, RNA interference, and transcription factor binding have all been shown to play a role in limiting LINE-1 expression and restricting retrotransposition [29,30,31,32,33,34,35,36]. Many of these mechanisms are disrupted in cancer cells, allowing for the re-expression and mobilization of LINE-1 [37,38,39]. LINE-1 protein ORF1p has been observed in approximately 47% of tumors, and has been a proposed indicator of aggressive disease in some cancers [37, 40, 41]. New LINE-1 insertions have also been detected in around 53% of tumors studied. In prostate cancer, 60% of tumors contained at least one new LINE-1 insertion, and the rate of retrotransposition was accelerated in metastatic disease [38]. However, this varies among cancers because in clear cell renal cell carcinoma (ccRCC), no new insertions were detected in tumor samples assessed [42]. This variation in LINE-1 expression and retrotransposition between types of cancers suggests possible cell-type specific regulation of LINE-1. While LINE-1 insertions are frequently found in introns and non-coding regions, new exonic insertions have also been detected in tumor suppressor genes, including APC in colorectal cancer [33, 43]. In addition to directly disrupting a gene, new insertions can alter the regulatory landscape of the genome by inducing new patterns of methylation [44,45,46].
The potential for LINE-1 to alter gene expression and drive genomic instability suggests that it may promote cancer. Better understanding of LINE-1 transcriptional regulation will provide insight into its dysregulation and activity in cancer cells. To date, Suv39h H3K9me3, the HUSH complex, SETDB1, and KAP1 were all shown to regulate LINE-1 elements in embryonic cells [47,48,49,50]. Additional studies have identified Myc, CTCF, YY1 and RUNX3 binding sites on the LINE-1 5’UTR through motif analysis [32, 51,52,53]. Further functional analysis of these transcription factors have revealed roles in transcriptional regulation and transcription initiation [32, 52]. Here, we have optimized a unique CRISPR C-BERST model to conduct an unbiased study identifying transcriptional regulators of active LINE-1 (L1Hs) in cancer cells. While this technique can identify traditional transcription factors, it can also identify proteins that are not directly bound to DNA but also play role in transcriptional regulation.
Results
Targeting the LINE-1 5’UTR promoter with dCas9 C-BERST
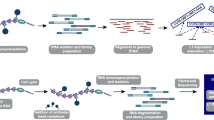
To better understand LINE-1 transcriptional regulation in cancer cells, we utilized the dCas9 C-BERST (dCas9–APEX2 Biotinylation at genomic Elements by Restricted Spatial Tagging) method to map regulatory proteins bound to the LINE-1 promoter [54]. C-BERST utilizes a nuclease deficient Cas9 (dCas9) fused to the ascorbate peroxidase APEX2. When expressed, dCas9-APEX2 is directed to specific DNA loci using guide RNAs (gRNA). Upon treatment with biotin-phenol and hydrogen peroxide, APEX2 generates biotin-phenoxyl radicals which covalently biotin-label proteins within an ~ 20 nm radius (Fig. 1A) [54]. The LINE-1 promoter is located within its 5’UTR and is transcribed as part of LINE-1 mRNA to preserve it during retrotransposition [11]. To map regulatory proteins directing LINE-1 transcription in cancer cells we directed gRNAs to the LINE-1 5’UTR promoter region.
Utilizing the C-BERST method to target LINE-1 5’UTR. A An APEX2 tagged dCas9 is recruited to the LINE-1 5’UTR promoter using L1Hs specific gRNAs. Cells are incubated with biotin-phenol for 30 min and treated with hydrogen peroxide for 1 min, triggering the biotinylation of proteins located at the LINE-1 5’UTR promoter. Biotinylated proteins are enriched through streptavidin immunoprecipitation and identified through mass spectrometry. Figure and method based on Gao et al. [54]. B Alignment of L1PA2-L1PA7 to L1Hs. Blue lines designate correct alignment to L1Hs, cream lines designate mismatch alignment to L1Hs, and arrowheads mark additional misaligned sequence missing from L1Hs. Eight guide RNAs (gRNA) were designed to target the 5’UTR of LINE-1. Preference went to guides that aligned well with L1Hs, but lacked alignment to older LINE-1 sequences (L1PA2-L1PA7). C Chromatin immunoprecipitation (ChIP) of dCas9 was performed in cells expressing each of the eight LINE-1 5’UTR gRNAs, as well as a non-targeting control (NS). ChIP reads were aligned to LINE-1 L1Hs, as well as older L1PA2-L1PA7, using MapRRCon analysis software [32]. Red line denotes the target location of each gRNA. Representative data shown for gRNA 3, 4, 7 and NS. Complete data set for all gRNAs can be found in Supplemental Figure 1
To selectively direct dCas9 to active, retrotransposition competent LINE-1 sequences, we designed eight gRNAs that specifically targeted the L1Hs 5’UTR. gRNA design targeted regions of the 5’UTR that aligned poorly to older LINE-1 sequences, L1PA2-L1PA7 (Fig. 1B). Next, we conducted chromatin immunoprecipitation (ChIP) of dCas9 in the presence of each 5’UTR gRNAs and one non-targeting gRNA control (NS). Using the LINE-1 specific software MapRRCon [32], we aligned Cas9 ChIP reads to L1Hs, as well as to the older LINE-1 elements, L1PA2-L1PA7. As anticipated, all eight gRNAs aligned to the targeted region in the 5’UTR of L1Hs (Supplementary Figure S1). When we aligned ChIP reads to the older, nonfunctional LINE-1 sequences (L1PA2-L1PA7), gRNA 4, gRNA 7, and gRNA 8, were enriched for L1Hs binding and began to lose alignment quality to older sequences beginning with L1PA7. Guides 4, 7 and 8 had lost almost all alignment to L1PA3-7, making them the least likely to target ancestral LINE-1 (Fig. 1C). All other guides showed alignment with L1PA2 and L1PA3, as shown with gRNA 3, but lost significant peaks in the 5’UTR as they were aligned to older LINE-1 sequences (Fig. 1C, Supplementary Figure S1). gRNA 4 and gRNA 7 were chosen for the C-BERST assay due to their high level of specificity for younger, active, L1Hs sequences, and minimal recruitment to the older L1PA2-L1PA7 sequences.
Identification of LINE-1 5’UTR localized proteins through dCAS9-APEX2 biotinylation
The two cell lines we used to identify LINE-1 5’UTR bound proteins were LNCaP and E006AA-hT. LNCaP cells are an androgen dependent prostate cancer cell line that has been shown to express LINE-1 ORF1 protein and mRNA (Fig. 2A and B). E006AA-hT cells express no detectable LINE-1 ORF1 protein [55], and have very low levels of LINE-1 mRNA (Fig. 2A and B). While first thought to be a prostate cancer cell line, E006AA-hT cells were later found to be a clone of renal cell carcinoma cell line 786‐O [56]. The variation of LINE-1 expression in these cell lines provides a compelling model to better understand LINE-1 transcriptional activation and repression in cancer cells. For example, proteins bound to the LINE-1 5’UTR in E006AA-hT cells may have suppressive activity since the cells do not express LINE-1.
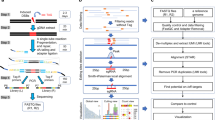
C-BERST identification of 5’UTR biotinylated transcription factors. A Western blot of LINE-1 ORF1p expression in whole cell lysates of E006AA-hT and LNCaP cells. (Tubulin loading control). B Relative LINE-1 mRNA levels as assessed by qPCR. C Western blot of C-BERST streptavidin immunoprecipitation. Cells expressing gRNA-4, gRNA-7, or gRNA-NS were harvested, and biotinylated proteins were collected by streptavidin immunoprecipitation. A no hydrogen peroxide control (No H2O2) was included in the harvest to eliminate endogenously biotinylated proteins. Input, flow through, and immunoprecipitation (IP) were blotted with streptavidin HRP. D Proteins identified with C-BERST that were enriched at least 1.5 × in both LINE-1 targeting gRNAs (gRNA-4, gRNA-7) when compared to the non-targeting control (gRNA NS). Transcription factors have been highlighted in red. E Complete list of transcription factors enriched at least 1.5 × in both LINE-1 targeting gRNAs (gRNA-4, gRNA-7) when compared to non-targeting control (gRNA NS). Proteins previously identified to regulate LINE-1 are highlighted in green. Blue squares denote ENCODE ChIP data availability. Red squares denote peak on specified LINE-1 location by MapRRCon analysis
Each cell line was stably transfected with dCas9-APEX2 and a gRNA (gRNA 4, gRNA 7, or gRNA NS). As previously optimized, cells were sorted to select for low dCas9 (mCherry) expression in order to reduce background [54]. dCas9-APEX2 was induced with doxycycline treatment for 21 h, cells were incubated with biotin-phenol for 30 min, and treated with hydrogen peroxide for 1 min. After quenching the hydrogen peroxide and isolating nuclei, biotinylated proteins were collected through streptavidin immunoprecipitation and proteins were identified by mass spectroscopy (Fig. 2C and D). Our screen revealed 22 transcription factors in LNCaP cells (356 total enriched proteins), and 24 transcription factors in E006AA-hT cells (149 total enriched proteins), that were enriched at least 1.5 × above gRNA NS in both LINE-1 specific guides (gRNA 4, gRNA 7) (Fig. 2D and E). In both screens, we identified proteins previously shown to regulate LINE-1 expression, including YY1, PPHLN1, and CTCF in LNCaP cells, and DNMT1 in E006AA-hT cells (Fig. 2E) [32, 48, 52, 53, 57].
Validating transcription factors with MapRRCon
To assess the presence of enriched transcription factors on the LINE-1 5’UTR we analyzed available ENCODE ChIP data with MapRRCon software. For LNCaP cells, we analyzed 15 transcription factors from our C-BERST screen that had available ENCODE ChIP data and found that 12 of these proteins (80%) showed peaks on LINE-1 with 9 mapping to the 5’UTR (60%) (Figs. 2E and 3B). Similarly, we also analyzed 12 transcription factors enriched in the E006AA-hT C-BERST assay and found that 7 (58.3%) contained peaks on LINE-1 with 5 mapping to the 5’UTR (41.6%) (Figs. 2E and 3A). In both cell lines there were transcription factors that showed more than one peak on full length LINE-1, including RCOR2 (Fig. 3). Interestingly, ENCODE ChIP data was collected from multiple cell types and was analyzed by MapRRCon for peaks on LINE-1. Many of the identified transcription factors had peaks in multiple cell types, suggesting a broad role in regulating LINE-1 transcription (Fig. 3), while others, such as BCL3, showed clear differences between cell types (Supplemental Figure S2).
MapRRCon ChIP plots of C-BERST identified proteins. MapRRCon analysis of C-BERST enriched proteins in A E006AA-hT and B LNCaP cells. Each plot is an analysis of available ENCODE ChIP data, in multiple cell lines (listed on the right side of each plot). Position of observed peaks is noted by the full length L1Hs diagram along the bottom of the figure. Additional MapRRCon plots can be found in Supplemental Figure 2
C-BERST reveals novel regulators of LINE-1 mRNA expression
The top enriched protein in our E006AA-hT C-BERST screen was DUSP1, a dual specificity protein phosphatase known to inhibit MAPK [58]. In order to test the effect of DUSP1 on LINE-1 expression, we conducted a knockdown of DUSP1 with shRNA in E0006AA-hT cells. We evaluated LINE-1 mRNA levels by qPCR and found a 1.6 fold increase of LINE-1 transcript levels upon DUSP1 knockdown (Fig. 4A). Next, we used a DUSP1 inhibitor, BCI, and assessed its effect on LINE-1 transcript levels by qPCR. Again, we saw a 1.5–1.6 fold increase in LINE-1 transcript levels upon treatment (Fig. 4B). DUSP1 was only found in the E006AA-hT C-BERST screen, suggesting that it is inhibiting LINE-1 expression; to test this hypothesis, we examined its effect on LINE-1 transcript levels in LNCaP cells. Upon over expression of DUSP1 in LNCaP cells, we observed a 45% decrease in LINE-1 transcript levels (Fig. 4C) supporting our hypothesis. Overexpression of DUSP1 in E006AA-hT cells resulted in no change in LINE-1 levels, likely due to DUSP1 saturation in these cells. We also observed high DUSP1 mRNA levels in PC3 cells, a prostate cancer cell line shown to have low levels of LINE-1 expression (Fig. 4D) [55]. To examine whether DUSP1 was playing a role in regulating LINE-1 in PC3 cells, we knocked down DUSP1 with siRNA and evaluated LINE-1 mRNA and ORF1 protein levels (Fig. 4E). After knockdown, we observed an increase in both LINE-1 mRNA and ORF1p protein levels. ORF1p protein levels increased by 2.0 and 2.4 fold compared to siScramble. Together, our results show DUSP1 plays a role in suppression of LINE-1 transcription in cancer cells.
DUSP1 modulates LINE-1 mRNA expression. A qPCR of LINE-1 mRNA levels in E006AA-hT cells expressing shRNA to DUSP1 or non-targeting control (NS). B qPCR of relative LINE-1 levels in E006AA-hT cells that were treated with vehicle (DMSO) or DUSP1 inhibitor BCI. C qPCR of relative LINE-1 mRNA levels in E006AA-hT or LNCaP cells overexpressing vector only (VO) or DUSP1. D qPCR of DUSP1 mRNA levels in panel of prostate cancer cell lines (including E006AA-hT renal cell carcinoma line). E qPCR of relative LINE-1 mRNA levels (left) and western blots of LINE-1 ORF1p, DUSP1, and hsp90 (loading control) protein levels (right) in PC3 cells that were treated with siScramble or siDUSP1 siRNA. ORF1 protein levels relative to hsp90 were quantified, siScramble was set to 1
Discussion
The dysregulation of LINE-1 in cancers and its potential to perturb genomic integrity underscores the importance of understanding LINE-1 transcriptional regulation. However, the abundance of ancestral LINE-1 sequences in the human genome compounds the challenge of studying this repetitive element. Here, we utilized chromatin immunoprecipitation and LINE-1 specific analysis software, MapRRCon, to mitigate this challenge and assess the L1Hs specificity of eight CRISPR Cas9 gRNAs. Our analysis revealed three gRNAs, spanning a 47 bp section of the 5’UTR, that showed high specificity to L1Hs and low recruitment to L1PA2-L1PA7. These three gRNAs differed from the L1PA2 consensus sequence by 1 (gRNA4), or 2 (gRNA7, gRNA8) base pairs, and differed from L1PA3 by 2 (gRNA4, gRNA8) or 3 (gRNA7) base pairs. While these differences between our gRNAs and older LINE-1 sequences were subtle, our ChIP analysis shows that they disrupted gRNA recruitment to L1PA2-L1PA7. Out of these three gRNAs, we chose to use gRNA 4 and gRNA7 for our C-BERST assay. While gRNA 8 also showed high specificity to L1Hs, it did not fully align with an active L1Hs sequence that we previously identified to be expressed in LNCaP cells (Xp22.2(2)) [21]. Since Xp22.2(2) was one of the highest LINE-1 loci expressed in LNCaP cells, we decided to exclude it as a guide. However, gRNA 8 may be used as a viable LINE-1 gRNA in alternative cell types. The analysis of these gRNAs not only set up a platform for targeting L1Hs with C-BERST, it also provides insight for additional CRISPR applications used to target LINE-1 L1Hs.
We preformed C-BERST in two different cell types, LNCaP and E006AA-hT, in order to assess different mechanisms of LINE-1 regulation. In LNCaPs, we have previously shown that many different LINE-1 loci are being actively expressed, with notable ORF1p levels observed [21, 55]. While LNCaP cells express a substantial number of LINE-1 loci, they contained a range of expression from highly expressed LINE-1 loci to non-detectable loci as analyzed by ORF1p RNA immunoprecipitation [21]. On the other hand, E006AA-hT cells have very low LINE-1 mRNA expression with no detectable ORF1p expression (Fig. 2A and B). In LNCaP cells we identified 356 proteins that were enriched at the 5’UTR in both gRNAs, 22 of which were transcription factors. In E006AA-hT cells, we only observed 149 enriched proteins, including 24 transcription factors. The discrepancy in total number of enriched proteins in each cell line may be due to the variation in LINE-1 expression in LNCaP cells. Different LINE-1 loci may have a variety of protein complexes recruited for either LINE-1 activation or repression, yielding a higher number of enriched proteins. This discrepancy may also be cell type specific, or possibly due to higher background in LNCaP cells. Among the transcription factors identified only one, ZBTB33, was enriched in both LNCaP and E006AA-hT cell lines. These results suggest a dramatic difference in LINE-1 regulation between cell and/or cancer type.
The C-BERST method was developed to identify proteins bound or localized to specific loci. Our screen revealed a number of proteins that have previously been shown to regulate LINE-1 expression, as well as identified new putative LINE-1 regulators. Transcription factors YY1 and CTCF were both enriched at the 5’UTR in LNCaP cells and have previously been shown to bind to the LINE-1 5’UTR and modulate transcription [32, 52, 53]. PPHLN1, a component of the HUSH complex, was highly enriched in our LNCaP screen, and has also been shown to inhibit LINE-1 expression [48, 59]. The HUSH complex maintains H3K9me3 through the recruitment of SETDB1, another protein shown to regulate LINE-1 expression and found to be enriched in our assay [60, 61]. However, other than PPHLN1, no other components of the HUSH complex were identified as enriched, perhaps because LINE-1 is not repressed at many LINE-1 loci in LNCaP cells. Additionally, DNMT1, a DNA methyltransferase that has previously been shown to repress young LINE-1 elements, was enriched at the 5’UTR in E006AA-hT cells [57]. In addition to these previously identified proteins, our MapRRCon analysis identified five proteins in E006AA-hT cells and nine in LNCaP cells that had confirmed ChIP peaks in the LINE-1 5’UTR. Additional proteins were shown to have peaks in ORF2 and in the 3’UTR in our MapRRCon analysis. It is possible that these proteins were enriched due to secondary chromatin structure at the LINE-1 loci, however, further analysis is needed to determine their potential role in LINE-1 regulation.
Dual Specificity Phosphatase 1, DUSP1, was the most highly enriched protein in E005AA-hT cells. DUSP1 inactivates MAPK through dephosphorylation of threonine/tyrosine, including p38 MAPK, JNKs and ERKs [58, 62]. We speculate that DUSP1 may dephosphorylate MAPKs and thereby alter downstream transcription factor activity in protein complexes regulating the L1HS 5’UTR. In early prostate and bladder cancers, DUSP1 is expressed at higher levels, however, as histological grade progresses, DUSP1 levels decrease. Our results show that DUSP1 consistently contributes to LINE-1 repression in E006AA-hT cells and PC3 cells. Interestingly, high DUSP1 expression was observed in cell lines with lower LINE-1 expression (Fig. 4D) [55]. While our C-BERST results strongly suggest it is localized around the 5’UTR, further analysis is needed to explore DUSP1 substrates instrumental in LINE-1 regulation. Since we required proteins to be enriched in both guides, it is possible that our stringency eliminated important cofactors and DUSP1 substrates. Our application of C-BERST enabled us to identify proteins both directly bound to the LINE-1 5’UTR (ZBTB33, ERG1, GATAD1 etc.), as well as transient regulators that may have been missed in traditional sequence and ChIP analysis (DUSP1). Overall, our LINE-1 optimized C-BERST assay enables the identification of cell type specific LINE-1 transcriptional regulators.
Conclusions
The abundance of ancestral LINE-1 sequences in the genome presents a significant challenge to studying active L1Hs regulation. In our study, we identified three CRISPR Cas9 gRNAs that specifically target active L1Hs, with minimal binding to older LINE-1 sequences. We also utilized these gRNAs in the restricted spatial tagging method, C-BERST, to identify proteins localized to the LINE-1 5’UTR promoter. Our application of the C-BERST method identified both known and novel LINE-1 transcriptional regulators, including the dual specificity phosphatase, DUSP1, in cancer cells. Our optimization of the C-BERST method to specifically target the L1Hs promoter has created a tool that can be used to better understand the regulation of LINE-1 expression in cancer cells.
Materials and methods
Cell culture
E006AA-hT (CRL-3277) and LNCaP (CRL-1740) cell lines were purchased from the ATCC. E006AA-hT cells were maintained in DMEM supplemented with 10% FBS. LNCaP cells were maintained in RPMI 1640 supplemented with 10% FBS. Cells were assessed regularly for mycoplasma contamination.
siRNA knockdown
Human DUSP1 siRNA SMARTpool (#L-003484–02-005) and non-targeting control pool (#D-001810–10-05) was purchased from Dharmacon. PC3 cells (2.5 × 105 cells per well) were seeded on 6 well plates. PC3 cells were transfected with 25 and 50 nM siRNAs using Lipofectamine RNAiMAX reagent (Life Technologies) according to the manufacturer’s instructions. Transfections were performed on two consecutive days. RNA and protein were collected 72 h after the first transfection. Whole cell lysates were harvested in RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 1% NP-40, 0.1% SDS, 10 mM EDTA, 10 μg/mL aprotonin and leuptin, 1 mM PMSF, and 1 mM Na3VO4) and protein concentration was quantified using a Bradford Assay.
shRNA knockdown
DUSP1 and scramble shRNA were cloned into a pTRIPZ backbone. pTRIPZ plasmid and viral packaging plasmids were transfected into HEK 293 T cells with Lipofectamine Reagent (Thermo Fisher 18324012) (2 μg pMD2G, 3 μg psPAX2, and 5 μg pTRIPZ) and virus was collected and filtered after 48 h. Viral supernatant (4 mL) was supplemented with fresh media (2 mL) and Polybrene (8 μg/mL) and incubated with E006AA-hT cells for 4 h. After 48 h, cells were treated with puromycin (1 μg/mL). Once selected, shRNA expression was induced with 1 μg/mL doxycycline for 48 h.
sgRNA creation and design
sgRNAs were designed using the MIT guide RNA design tool (CRISPR.MIT.edu). sgRNAs with at least one mismatch to older LINE-1 (L1PA2-L1PA7) sequences were prioritized. Once sequences were selected, sgRNAs constructs were cloned by into the pEJS614_pTetR-P2A-BFPnls/sgNS by replacing sgNS sequence with LINE-1 targeting guide sequence. Guide sequences can be found in Supplementary Table 2.
DUSP1 overexpression
DUSP1 sequence was amplified from a DUSP1 GenScript plasmid (OHu10841D) and cloned into a pCW57-MCS1-2A-MCS2 backbone (Addgene #71782). Overexpression plasmid and viral packaging plasmids were transfected into HEK 293 T cells with Lipofectamine Reagent (Thermo Fisher 18324012) (2 μg VSV-G, 3 μg gag-pol, and 5 μg Overexpression plasmid) and virus was collected and filtered after 48 h. Viral supernatant (4 mL) was supplemented with fresh media (2 mL) and Polybrene (8 μg/mL) and incubated with E006AA-hT or LNCaP cells for 4 h. After 48 h, cells were treated with puromycin (1 μg/mL). Once selected, DUSP1 expression was induced with 1 μg/mL doxycycline for 48 h.
BCI treatment
E006AA-hT cells were seeded in a 6-well plate and incubated overnight at 37 °C. Cells were then treated with DMSO or BCI (Axon Medchem #2178) for 3 h at 37 °C. RNA was collected with the Qiagen RNeasy Plus Mini Kit (74134) as described below and assessed by qPCR.
RNA isolation and qPCR
RNA was isolated from cells using the Qiagen RNeasy Plus Mini Kit (74134) and contaminating DNA was digested using a Turbo DNA-free DNase digestion (Thermo Fisher Scientific AM1907) according to manufacturer’s protocol. cDNA was made using the Verso cDNA kit (Thermo Scientific- AB1453A). qPCR was conducted using SYBR Green Master Mix (Life Technologies 4344463) and relative mRNA levels were calculated using ΔΔCT. RPL19 was used as an internal control for normalization. Primer sequences can be found in Supplementary Table 2. qPCR LINE-1 primers have been previously published [63].
Chromatin immunoprecipitation
E006AA-hT and LNCaP cells (~ 20 × 106) stably expressing dSpyCas9-mCherry-APEX2 and gRNA were grown to 80% confluency. Cells were treated with 250 nM Sheild1 (Clontech) and 2 μg/mL doxycycline for 21 h to induce dCas9 expression. Cells were crosslinked with formaldehyde (1% formaldehyde in PBS) at room temperature for 10 min, and quenched with 1 mL of 2.5 M glycine, gently shaking for 5 min. Cells were washed with PBS and pelleted at 425xg for 5 min at 4 °C and resuspended in Farnham lysis buffer (5 mM PIPES pH 8.0, 85 mM KCL, 0.5% NP-40, Halt protease inhibitor (Thermo Fisher-87786)). Suspension was re-pelleted and flash frozen in liquid nitrogen. Pellets were resuspended in 1 mL Farnham lysis buffer with Halt protease inhibitor, passed through a 25 gauge syringe 15 times, and spun at 425xg for 5 min at 4 °C. Pellets were resuspended in RIPA buffer (1 × PBS, 1%NP-40, 0.5% Na-deoxycholate, 0.1% SDS, Halt protease inhibitor) and passed through a 25 gauge syringe 20 times. Lysates were sonicated in a Diagenode Bioruptor for 30 min, 30 s on, 30 s off at 2 °C and spun at 20800xg for 10 min. Sheared DNA was incubated with 4 μg mCherry antibody (Thermo PA5-34,974) overnight at 4 °C and incubated with 50μL protein A/G beads for 3 h at 4 °C. Beads were washed 5 × with LiCl wash buffer (100 mM Tris pH 7.5, 500 mM LiCl, 1% NP-40, 1% Na-deoxycholate) for 3 min, and once with TE buffer (10 mM tris–HCl pH7.5, 0.1 mM Na2EDTA) for 1 min. Beads were resuspended in Proteinase K/SDS solution (0.5% SDS, 0.2 mg/mL Proteinase K, 1X TE) and incubated at 55 °C for 3 h and 65 °C overnight to reverse crosslinks. Samples were placed on magnetic strip to collect supernatant. 600μL of PB and 4μL of RNaseA (17500u/mL) were added to the samples, and samples were purified using the Qiagen PCR purification kit (28104). Sample was eluted twice with 35μL of 10 mM Tris pH 8. Illumina libraries were generated using the NEB Next DNA Library Prep Ultra II kit (E7645S) according to manufacturer’s protocol. Libraries were sequenced on an Illumina NextSeq 500. Reads were demultiplexed with Illumina bcl2fastq v2.20 requiring a perfect match to indexing BC sequences.
Cell sorting
LNCaP and E006AA-hT cells were treated with doxycycline (2 μg/mL) and Sheild1 (250 nM) for 21 h prior to sorting. Cells were sorted using the SONY SY3200 parallel sorter (SONY Biotechnology, San Jose, CA), using a 100 µm orifice nozzle and system pressure of approximately 25 psi. Double positive cells for mCherry and BFP were purified as previously described [54].
C-BERST assay
Biotinylation: Seven 15 cm plates of E006AA-hT or LNCaP cells (~ 6 × 107) expressing a gRNA and dCas9-APEX2 were treated with 2 μg/mL doxycycline and 250 nM Sheild 1 for 21 h. Cells were incubated for 30 min with biotin-phenol (500 μM) at 37 °C, and 1 mM H2O2 was then added to cells for 1 min at room temperature. To stop the biotinylation reaction, quencher solution (5 mM trolox, 10 mM sodium ascorbate, and 10 mM sodium azide) was added and cells were placed on ice. Three additional washes with quencher solution were performed, followed by two washes with PBS.
Nuclear Isolation: Cells were scraped from plates and centrifuged at 300 × g for 5 min at 4 °C. Pellet was resuspended in 7.5 nuclei isolation buffer (10 mM PIPES pH 7.4, 0.1% NP-40, 10 mM KCl, 2 mM MgCl2, 1 mM DTT and Halt protease inhibitor). Cells were incubated on ice for 10 min and ruptured using a Dounce homogenizer (~ 20x). Cells were further incubated on ice for 20 min and homogenization was repeated. Lysate was gently added to a sucrose cushion that contained 20 mL of 30% sucrose and 3.5 mL 10% sucrose (10 mM PIPES pH 7.4, 10 mM KCl, 2 mM MgCl2, 30% or 10% sucrose, and 1 mM DTT). Sucrose cushion and cell lysate was spun at 1000 × g for 15 min. Supernatant was removed and nuclei (pellet) was resuspended in 800μL PBS. Suspension was spun at 1500 × g for 5 min at 4 °C. 500μL RIPA lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.125% SDS, 0.125% sodium deoxycholate, 1% Triton X-100) was added and samples were incubated at 4 °C. Lysates were sonicated in a Diagenode Bioruptor for 15 min (30 s on, 30 s off), and centrifuged for 10 min at 15,800 × g, 4 °C. Protein concentrations were measured using a Bradford Assay and samples were normalized.
Immunoprecipitation: MyOne Streptavidin T1 Dynabeads (Thermo Fisher 65,601) (400μL) were added to each sample and incubated at 4 °C overnight. Beads were washed with RIPA (twice), 1 M KCl, 0.1 M Na2CO3, 2 M urea in 10 mM Tris–HCl pH 8.0, and again with RIPA (twice). After washes, beads were processed for mass spectroscopy (see On-beads digestion of streptavidin-bound proteins). Protocol was based off of the previously described C-BERST technique [54].
Western blot/streptavidin blot
Cell lysates were boiled at 98 °C for 5 min in SDS loading buffer. Samples were resolved by SDS-PAGE on polyacrylamide gels, and transferred to PVDF using the BioRad Trans-Blot Turbo Transfer System. Blots were blocked in 5% BSA in TBS and probed with streptavidin-HRP (Thermo Fisher SA10001), ORF1p (Millipore MABC1152), DUSP1 (Cell Signaling 48625), or HSP90 (BD Biosciences 610419). Western blots were developed using BioRad Clarity Western ECL Substrate (1705060S) and visualized on an iBrightCL1000 Imager. Protein bands were quantified using ImageJ [64].
On-beads digestion of streptavidin-bound proteins
Streptavidin beads were washed twice with 1 mL 50 mM NH4HCO3 to exchange the buffer. Washed beads were then resuspended in 50 μl 50 mM NH4HCO3 containing 20 ng/μl trypsin/Lys-C (Promega) followed by overnight incubation at 37 °C with vigorous mixing in a thermoshaker (Eppendorf). After incubation beads were pelleted and supernatants were transferred to new tubes. Samples were acidified by adding 5 μl 20% heptafluorobutyric acid, incubated at room temperature for 5 min and clarified by 5-min centrifugation at 16000 g. Peptides from clarified samples were desalted using C18 spin tips (Thermo Scientific) according to manufacturer’s instructions. Desalted peptides were dried under vacuum and redissolved in 0.1% formic acid prior to LC–MS analysis. Peptide concentration was measured at 205 nm on Nanodrop One (Thermo Scientific).
LC–MS analysis
Peptides were analyzed by LC–MS on Orbitrap Fusion Lumos mass spectrometer coupled with Dionex Ultimate 3000 UHPLC. During each run, 0.5–2 μg of peptides from individual samples were injected and resolved on 50-cm long EASY-Spray column (Thermo Scientific) by 90-min long linear gradient of 4–40% acetonitrile in 0.1% formic acids at flowrate 0.25 μl/min. The method for data-dependent acquisition was based on published protocol [65] with exception that each cycle was set to last for 2 s instead of 3 s.
Peptides identification and label-free quantitation was done in the Proteome Discoverer 2.1. The protein database for Sequest HT search engine included human proteome downloaded from UniProt (www.uniprot.org) and amino acid sequence of streptavidin from Streptomyces avidinii. Parameters were set to search for peptides of at least 5 amino acids long, containing at most 2 missed trypsin cleavages. Dynamic modifications were set to include: phosphorylation of serine, threonine or tyrosine, acetylation of protein N-terminus, mono- and dimethylation of lysine and arginine. MS1-based label-free quantitation was done using the “Precursor Ions Area Detector” module in Proteome Discoverer. Samples were first normalized by intensity of streptavidin detected. Samples with an area value of 0 were replaced with the lowest MS1 intensity detected. Next, H2O2 only (endogenous biotinylation) values were subtracted from the + H2O2 samples (gRNA NS, gRNA 4 and gRNA7). Mean was calculated from replicates and targeted guides (gRNA 4 and gRNA 7) were divided by gRNA NS values for each protein detected. Proteins with at least 1.5 × greater than gRNA NS control were considered enriched and included in further analysis.
Availability of data and materials
Reagents will be available upon request.
Abbreviations
- LINE-1:
-
Long Interspersed Element-1
- C-BERST:
-
DCas9-APEX2 Biotinylation at genomic Elements by Restricted Spatial Tagging
- DUSP1:
-
Dual specificity protein phosphatase 1
References
Konkel MK, Walker JA, Batzer MA. LINEs and SINEs of primate evolution. Evol Anthropol. 2010;19(6):236–49.
Boeke JD, et al. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500.
Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921.
Szak ST, et al. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3(10):research0052.
Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. EMBO J. 1984;3(8):1753–9.
Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100(9):5280–5.
Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–75.
Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141(7):1159–70.
Scott AF, et al. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1(2):113–25.
Dombroski BA, et al. Isolation of an active human transposable element. Science. 1991;254(5039):1805–8.
Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10(12):6718–29.
Criscione SW, et al. Genome-wide characterization of human L1 antisense promoter-driven transcripts. BMC Genomics. 2016;17:463.
Denli AM, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163(3):583–93.
Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21(6):1973–85.
Jacobs FM, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–5.
Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16(1):78–87.
Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–27.
Martin SL, et al. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci U S A. 2003;100(24):13815–20.
Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21(2):467–75.
Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci U S A. 1997;94(19):10155–60.
Briggs EM, et al. RIP-seq reveals LINE-1 ORF1p association with p-body enriched mRNAs. Mob DNA. 2021;12(1):5.
Feng Q, et al. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–16.
Mathias SL, et al. Reverse transcriptase encoded by a human transposable element. Science. 1991;254(5039):1808–10.
Mita P, et al. LINE-1 protein localization and functional dynamics during the cell cycle. Elife. 2018;7:e30058.
Cost GJ, et al. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21(21):5899–910.
Luan DD, et al. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595–605.
Idica A, et al. MicroRNA miR-128 represses LINE-1 (L1) retrotransposition by down-regulating the nuclear import factor TNPO1. J Biol Chem. 2017;292(50):20494–508.
Kubo S, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A. 2006;103(21):8036–41.
Chen L, et al. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics. 2012;7(7):758–71.
Goodier JL, Cheung LE, Kazazian HH Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8(10):e1002941.
Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–40.
Sun X, et al. Transcription factor profiling reveals molecular choreography and key regulators of human retrotransposon expression. Proc Natl Acad Sci U S A. 2018;115(24):E5526–35.
Scott EC, et al. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016;26(6):745–55.
Nguyen THM, et al. L1 retrotransposon heterogeneity in ovarian tumor cell evolution. Cell Rep. 2018;23(13):3730–40.
Ewing AD, et al. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol Cell. 2020;80(5):915-928.e5.
Grundy EE, Diab N, Chiappinelli KB. Transposable element regulation and expression in cancer. FEBS J. 2021. https://doi.org/10.1111/febs.15722
Rodic N, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184(5):1280–6.
Tubio JM, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345(6196):1251343.
Burns KH. Transposable elements in cancer. Nat Rev Cancer. 2017;17(7):415–24.
Harris CR, et al. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer. 2010;1(2):115–24.
Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331(6017):593–6.
Helman E, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24(7):1053–63.
Miki Y, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–5.
Beck CR, et al. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215.
Grandi FC, et al. Retrotransposition creates sloping shores: a graded influence of hypomethylated CpG islands on flanking CpG sites. Genome Res. 2015;25(8):1135–46.
Garcia-Perez JL, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466(7307):769–73.
Bulut-Karslioglu A, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014;55(2):277–90.
Robbez-Masson L, et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018;28(6):836–45.
Castro-Diaz N, et al. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28(13):1397–409.
Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25(6):594–607.
Yang N, et al. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31(16):4929–40.
Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32(13):3846–55.
Sanchez-Luque FJ, et al. LINE-1 evasion of epigenetic repression in humans. Mol Cell. 2019;75(3):590-604.e12.
Gao XD, et al. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat Methods. 2018;15(6):433–6.
Briggs EM, et al. Long interspersed nuclear element-1 expression and retrotransposition in prostate cancer cells. Mob DNA. 2018;9:1.
Koochekpour S, et al. Correction: Establishment and characterization of a primary androgen-responsive African-American prostate cancer cell line, E006AA. Prostate 2004;60(2):145-152. Prostate. 2019;79(7):815.
Jonsson ME, et al. Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat Commun. 2019;10(1):3182.
Franklin CC, Kraft AS. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J Biol Chem. 1997;272(27):16917–23.
Tunbak H, et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat Commun. 2020;11(1):5387.
Cuellar TL, et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J Cell Biol. 2017;216(11):3535–49.
Tchasovnikarova IA, et al. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348(6242):1481–5.
Chu Y, et al. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271(11):6497–501.
Goering W, Ribarska T, Schulz WA. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis. 2011;32(10):1484–92.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5.
Davis S, et al. Expanding proteome coverage with CHarge Ordered Parallel Ion aNalysis (CHOPIN) combined with broad specificity proteolysis. J Proteome Res. 2017;16(3):1288–99.
Acknowledgements
Erik Sontheimer generously provided the Cas9-Apex2 and sgRNA plasmids used for these experiments. (Addgene plasmid # 108649, # 108570). All sequencing was performed by the NYU Langone Institute for Systems Genetics. We would also like to thank Raven Luther and Megan Hogan for their technical expertise.
Conflict of interests
Jef Boeke is a Founder and Director of CDI Labs, Inc., a Founder of Neochromosome, Inc, a Founder and SAB member of ReOpen Diagnostics, and serves or served on the Scientific Advisory Board of the following: Sangamo, Inc., Modern Meadow, Inc., Sample6, Inc. and the Wyss Institute.
Funding
This work was supported by the National Institutes of Health Grants R01CA112226 (to S. K. L.), F31CA225053-01A1 (to E.M.B.), P01AG051449 (subcontract to J.D.B.), R21CA235521 to J.D.B, R01GM127267 (to E.N.), the Blavatnik Family Foundation (E.N.), and the Howard Hughes Medical Institute (E.N.).
Author information
Authors and Affiliations
Contributions
E.M.B, P.M., S.K.L., and J.D.B. designed the study. E.M.B, P.M., S.H. and Z.R.L. carried out experiments and analyzed data. X.S., P.M. and J.D.B. analyzed ENCODE ChIP data with MapRRCon software. N.V. performed the mass spectrometry. S.K.L., J.D.B. and E.N. provided supervisor oversight. E.M.B. wrote the manuscript. All authors provided input and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplemental Figure 1.
Complete ChIP assessment for CRISPR Cas9 gRNA localization to LINE-1 gRNA1-8 and gRNA-NS were expressed with dCas9 in cells. ChIP was performed with each guide and ChIP data was assessed with MapRRCon to quantify LINE-1 (L1Hs, L1PA2-L1PA7) localization of each gRNA. Position on full length LINE-1 along X-axis. Fold enrichment above input along Y-axis. Red dotted line marks gRNA target site.
Additional file 2: Supplemental Figure 2.
Additional MapRRCon ChIP Plots of C-BERST Enriched Transcription Factors. ChIP plots of enriched transcription factors with identified LINE-1 peaks.
Additional file 3: Supplemental Table 1.
C-BERST Enriched proteins in LNCaP and E006AA-hT cells. Complete list of proteins that were enriched at least 1.5 fold in gRNA 4 and gRNA 7 when compared to the non-targeting control (gRNA NS).
Additional file 4: Supplemental Table 2.
gRNA and qPCR Primer Sequences
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Briggs, E.M., Mita, P., Sun, X. et al. Unbiased proteomic mapping of the LINE-1 promoter using CRISPR Cas9. Mobile DNA 12, 21 (2021). https://doi.org/10.1186/s13100-021-00249-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13100-021-00249-9