Abstract
Background
Based on the results of existing observational studies, it can be found that the association between serum vitamin D levels and the risk of Sjogren’s syndrome (SS) in humans is still controversial. Based on this situation, this study aimed to assess the causal relationship between serum vitamin D levels and SS by using the Mendelian randomization (MR) approach.
Methods
In this study, genome-wide association studies (GWAS) summary statistics on serum vitamin D levels [sample size = 417,580 (UK Biobank)] and SS [sample size = 416,757 (cases = 2495, controls = 414,262) (FinnGen)] were used. The bi-directional MR analysis was then used to assess possible causal relationships. The major analysis method of MR was performed using inverse-variance weighted (IVW), supplemented by MR-Egger and the weighted median approaches. In addition, sensitivity analyses were used to ensure the stability of the results, including Cochran’s Q test, MR-PRESSO, MR-Egger intercept test, and the leave-one-out test.
Results
The MR suggested that no significant causal effects of serum 25(OH)D levels on SS risks were observed [odds ratio (OR) = 0.9824; 95% confidence interval (CI) = 0.7130 to 1.3538; P = 0.9137]. Similarly, no evidence supported the causal effects of SS on serum vitamin D levels (β: 0.0076, 95% CI: − 0.0031 to 0.0183; P = 0.1640).
Conclusion
This study found no obvious evidence that serum vitamin D level is causally associated with SS risks or vice versa. We call for larger sample size studies to further unravel the potential causal relationship and the exact mechanism.
Similar content being viewed by others
Introduction
Sjogren’s syndrome (SS) is a complex, heterogeneous systemic chronic autoimmune disorder commonly presenting with dry eyes and mouth [1,2,3]. SS is one of the most common autoimmune diseases with a prevalence of 0.1 to 4.8% in various populations, according to the strict definition of the American-European Consensus Criteria [4,5,6]. SS can cause damage to almost any organ or system causing a variety of complications, including immune thrombocytopenia, interstitial lung disease, autoimmune hepatitis, and lymphoma to name just a few [7], which places a tremendous financial burden on patients’ families and healthcare services [8]. In addition, SS can cause fatigue, depression, anxiety, and decreased physical performance, which in turn seriously affects the patient’s quality of life [5].
Vitamin D is a nutrient with multiple biological effects and its main form in serum is 25-hydroxyvitamin D [25(OH)D], which plays an important role in immune regulation [9, 10]. Currently, low levels of vitamin D due to lack of sunlight exposure or low dietary intake have been identified as a major risk factor for autoimmune diseases [11]. There have been several observational studies exploring the association between vitamin D and SS risk. Recently, two large cross-sectional studies including 107 and 176 SS patients from Turkey [12] and Europe [13] were conducted. The former study found no difference in vitamin D levels between cases and controls, while the latter reported lower levels of vitamin D levels in patients with SS. In addition, a cohort study demonstrated that vitamin D deficiency is common in SS patients [12], and a meta-analysis based on the observational studies also obtained the same results [14]. Based on the above, there are inconsistent results regarding the association between vitamin D levels and SS. Conclusions about causality cannot be drawn solely from the results in observational designs, possibly because of the limitations contained in the cohort and cross-sectional studies (limited sample size, different races, and other existing confounding factors and bias). Currently, it is uncertain whether the relationship between vitamin D and SS is causal, and whether it operates in one or both directions.
Mendelian randomization (MR) analysis is a useful epidemiological research strategy for assessing causal relationships. With the development and advancement of the Human Genome Project, MR analysis uses genetic variants as instrumental variables (IVs), which minimizes the limitations of observational studies and yields unconfounded information on the causal relationship between exposure and outcome through its specific analytical methods [15, 16]. IVs typically use single nucleotide polymorphisms (SNPs) obtained from genome-wide association studies (GWAS), which are DNA sequence polymorphisms induced by single nucleotide mutation within the genome [17]. According to the principle of independent classification (Mendel’s law of random allocation), genetic variants are randomly assigned during meiosis [18]; thus, they can be considered hereditary randomized controlled trials (RCTs) and may not be affected by residual confounding and reverse causality. Based on this, the study was to examine the causal association between serum vitamin D and SS, using the data from large-scale GWAS with the bi-directional MR design.
Materials and methods
Ethics
This study was reported according to STROBE-MR guidelines [19]. Data were collected from public databases. And there is no ethical approval necessary.
Study design
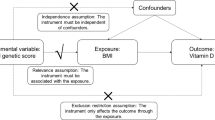
A bi-directional MR study was conducted to investigate the causal associations between serum vitamin D levels and SS [20]. SNPs are used as IVs for MR analysis to determine the causal effect of exposure variables [21, 22]. It is worth noting that MR analysis is subject to three assumptions [17]: (I) the IVs are closely related to exposure (“relevance”); (II) the IVs are independent of any potential confounding factor (“exchangeability”); (III) the IVs only affect outcome via the exposure (“exclusion restriction”). The framework is described in Fig. 1.
Data sources for serum 25(OH)D levels and SS
The summary data of serum 25(OH)D levels were obtained from the GWAS study at the UK Biobank [23], including phenotype, genotype, and clinical information on 417,580 individuals of European ancestry (age ranging from 40 to 69 years old) [24]. Serum 25(OH)D levels were quantified by chemiluminescent immunoassay in nanomoles per liter in blood samples collected from 2006 to 2013. Individuals with 25(OH)D levels out of the range of 10–375 nmol/L were excluded. The median, mean, and interquartile range of 25(OH)D levels were 47.9, 49.6, and 33.5 to 63.2 nmol/L, respectively.
Meanwhile, the summary data for SS was available from FinnGen [25] (https://www.finngen.fi/en/) with the phenocode “M13_SJOGREN” which consisted of 416,757 samples (2495 cases; 414,262 controls), and a total of 16,383,308 SNPs were genotyped. The information about each data source is provided in Table 1.
Selection of genetic variants as IVs
SNPs that were significantly associated with 25(OH)D levels and SS, respectively, were screened from GWAS data as preliminary IVs (P < 5 × 10−8 at genome-wide threshold). Meanwhile, linkage disequilibrium (LD) analysis was performed to ensure independence between SNPs (LD-r2 < 0.001 and clumping distance > 10,000 kb) [17]. Given that the main assumption of the MR analysis is that IVs can only affect the outcome through exposure, we manually eliminated SNPs related to confounders using PhenoScanner [26] (http://phenoscanner.medschl.cam.ac.uk/).
Testing instrument strength and statistical power
To minimize any possible weak IV bias, the strength of the IV was assessed using the F-statistic [27]. We use the following formula \(F=\frac{{R}^{2}\times (n-2)}{1-{R}^{2}}\) (n: sample size of the GWAS; R2: the proportion of explained variance of the IV) to calculate F-value [28]. A higher F-statistic corresponded to a smaller bias [27]. And if F > 10, it indicates that the study had sufficient strength [29]. Meanwhile, R2 was calculated using the formula: \({R}^{2}=2\times {\beta }^{2}\times (1-\mathrm{EAF})\times \mathrm{EAF}\) (β: estimate of the genetic effect of each SNP on iron status; EAF: effect allele frequency) [30]. The statistical power of MR analysis was calculated using an online tool at https://shiny.cnsgenomics.com/mRnd/ [31]. Briefly, power is calculated based on the sample size of GWAS, the proportion of cases, and the variance explained by genetic instruments for the exposure.
MR analysis
Before the MR analysis, the data were harmonized according to the previously described method [20], to correspond to the effect sizes of exposures and outcomes to the same effect alleles. The principal MR analysis was based on inverse-variance weighted (IVW) multiplicative random effects models [20]. The IVW method assumes that the MR assumptions are met or that all SNPs are valid. In addition, MR-Egger [32] and weighted median [33] were used in complementary analyses. MR-Egger analysis was conducted to assess whether the IVs have directional horizontal pleiotropic effects on the outcome [32]. The weighted median method can give a valid causal estimate when more than 50% of the information is derived from valid IVs [33]. However, the power of the weighted median and MR-Egger methods are limited compared to IVW, which tend to provide wider confidence intervals (CI) [20], and are therefore employed in this study only as complementary methods.
Sensitivity analyses
First, according to Cochran’s Q statistic, IV heterogeneity was determined by using the random effects model (P < 0.05) or fixed effects model (P > 0.05) [34]. Second, horizontal pleiotropy was examined by conducting the MR-Egger intercept test. Meanwhile, horizontal pleiotropy can be detected with the MR-Pleiotropy Residual Sum and Outlier methods (MR-PRESSO) based on both SNP-level and global heterogeneity estimates. To identify outlier variants, the outlier test compares expected and observed distributions of each variant. If any of the outlier variants are detected, they would be discarded to obtain an unbiased causal estimate from an outlier-corrected MR analysis [35]. Third, the leave-one-out method eliminated the included SNPs one by one and calculated the effect of the remaining IVs to evaluate whether the MR estimate was driven or biased by a single SNP, which was performed in the sensitivity analysis [18]. In addition, when vitamin D was used as the exposure and SS was used as the outcome, the measure of the effects was odds ratios (ORs) and its 95% CI; conversely, the effect measure was β and its 95% CI.
MR analyses were conducted using R version 4.2.1 with the “TwoSampleMR” [36] (version 0.5.6) and “MR-PRESSO” [37] (version 1.0) R packages. It was considered significant if the two-sided P-value was less than 0.05.
Results
Characteristics of the selected SNPs
SNPs strongly correlated with 25(OH)D were extracted as IVs in GWAS. The LD analysis was also performed (LD-r2 < 0.001, clumping distance > 10,000 kb). Meanwhile, SNPs for risk factors associated with SS (atopic dermatitis [38]: rs1038165, rs10454087, rs12123821, inflammation [39]: rs55814693) were searched (P < 1 × 10−5) through the PhenoScanner database. In addition, palindromic variants resulting in potential strand ambiguity were removed. Eventually, 81 SNPs were enrolled in the MR analysis of 25(OH)D on SS.
For the IVs of SS, we found that only a small number (n = 3) of SNPs were obtained when a strict P-value (P < 5 × 10−8) was taken for screening. To include more SNPs associated with SS, a more lenient threshold was used in this study (P < 5 × 10−7). After combining LD analysis and searching (P < 1 × 10−5) the PhenoScanner database for risk factors (basal metabolic rate [40]: rs3093958), four SNPs were finally obtained for the MR analysis of the causal association of SS on 25(OH)D.
The F-statistics for IVs were all over 10, suggesting IVs were generally considered to provide sufficient information for MR studies (Supplementary Tables 1 and 2).
Causal effects of serum 25(OH)D levels on SS risks
The results of this MR analysis are shown in Table 2 and Fig. 2, with a power of 0.32. The OR of serum 25(OH)D associated with SS for each of the three methods (IVW, weighted median, and MR-Egger) were 0.9824 (95% CI: 0.7130 to 1.3538, P = 0.9137), 1.1814 (95% CI: 0.7054 to 1.9786; P = 0.5263), and 0.8955 (95% CI: 0.5209 to 1.5394; P = 0.6908). Our results of the three methods all showed that genetically predicted levels of serum 25(OH)D were not significantly associated to the risk of SS (all P > 0.05).
Causal effects of SS on serum 25(OH)D levels
In Table 3 and Fig. 3, reverse MR results were presented with a low statistical power of 0.11. The β of SS associated with serum 25(OH)D for each of the three methods (IVW, weighted median, and MR-Egger) were 0.0076 (95% CI: − 0.0031 to 0.0183; P = 0.1640), 0.0114 (95% CI: − 0.0011 to 0.0239; P = 0.0773), and 0.0350 (95% CI: − 0.0030 to 0.0731; P = 0.2128). The results of the three methods all showed that genetically predicted SS were not significantly associated to levels of serum 25(OH)D (all P > 0.05).
Sensitivity analyses of MR
Our heterogeneity analysis showed that there was no heterogeneity between the data, as Cochran’s Q test was not statistically significant (Q = 71.3697, P = 0.7439). Moreover, in the MR-Egger test and the MR-PRESSO global test, no directional pleiotropy bias was evident (Tables 2 and 3). In addition, the leave-one-out analysis did not demonstrate any SNP outliers, which suggests that our results were stable (see Fig. 4).
Discussion
To our knowledge, this is the first bi-directional MR design to assess the causal effects between serum vitamin D and SS. However, our data provided evidence supporting no association between vitamin D and SS using the MR approach.
Our MR analysis contradicts some of the existing observational studies that have proposed a correlation between serum vitamin D levels and SS. In a cohort study of 107 SS patients (97 female and 10 male) and 74 healthy controls (64 female and 10 male), Erten et al. found that vitamin D deficiency was frequent in patients with SS, and female SS patients had the risk of vitamin D deficiency [12]. Moreover, the latest systematic review and meta-analysis revealed that SS patients have lower serum vitamin D levels than controls [standardized mean difference (SMD) = − 0.297; 95% CI: − 0.585 to − 0.01; P < 0.05] [14]. Although the correlation between serum vitamin D levels and SS was shown in the above studies, our MR study did not support a bi-directional causal association. This is consistent with the results of several other observational studies. Szodoray et al. found no statistical difference in serum vitamin D between 25 SS patients and 15 healthy individuals in a cross-sectional study [41]. In another cross-sectional survey, Agmon-Levin et al. similarly revealed no serological differences between 176 SS patients and 163 healthy individuals regarding vitamin D [13]. A possible explanation for the existence of such inconsistent results is that the previously observed associations between serum vitamin D levels and SS are coincidental or thwarted by an unknown confounder.
Moreover, a causal link between serum vitamin D levels and SS, or the converse, cannot be established in observational studies because SS is a systemic autoimmune disease accompanied by many complications, such as arthritis, rashes, pulmonary disease, renal or hepatic manifestations, central nervous system (CNS) involvement, and polyneuropathy [42]. Also, it is worth noting that vitamin D is a powerful immune modulator and that inflammation mutually regulates vitamin D metabolism [43, 44]. Therefore, we suspected the possibility that some inflammatory pathways would be shared between altered serum vitamin D levels and SS, leading to the conclusion in some observational studies that there is a correlation between the two phenotypes. Notably, the role of vitamin D has also been inconsistent in studies of its association with other autoimmune diseases. These include compulsory spondylitis [45], systemic lupus erythematosus [46], rheumatoid arthritis [47], and others. In sum, the underlying mechanism behind the relationship between vitamin D and SS is complex and worthy of further investigation. And due to the difficulty in observational epidemiological studies to eliminate the bias such as the reverse causal association of confounding factors, there are some limitations in etiological interpretation [48, 49].
In this study, the MR analysis satisfied three assumptions. For assumption I, the IVs are closely related to exposure. Eighty-one and four SNPs were selected in the GWAS, which was closely associated with vitamin D and SS, respectively, which verified assumption I. For assumption II, the IVs are independent of any potential confounding factor. LD between SNPs was assessed and screened, and we found that no SNPs were in LD with each other at an r2 > 0.05. For assumption III, the IVs only affect outcome via the exposure. The heterogeneity and sensitivity analyses have been conducted to detect and remove any potential pleiotropy, reassuring that our MR estimates are robust and reliable, with no perceptible bias from other sources of pleiotropy.
The strength of this study is the use of MR methods to minimize residual confounding and reverse causality in traditional observational studies. However, there are still some limitations that cannot be addressed at present. First, there should be no weak instrumental variable bias in this study because the F-statistics were all > 10; however, the low power may be caused by the low number of SNPs used as IVs. Meanwhile, this generated an impetus to perform MR studies on larger sample size populations. Second, the data obtained were GWAS summary data without specific personal information to the extent that subgroup analysis could not be performed. Third, because the population of European ancestry was used, we should be cautious about generalizing the findings to other populations. Meanwhile, the study needs to be repeated in future studies as the GWAS database continues to be improved and supplemented with sample size, sample information, etc.
Conclusions
In neither direction did we find causal evidence in support of a causal association between serum vitamin D levels and SS risk. However, multi-center, large-scale GWAS cohort studies are in development and may extend the IVs in this study. The association between vitamin D levels and SS needs to be re-evaluated in future studies.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- SS:
-
Sjogren’s syndrome
- MR:
-
Mendelian randomization
- GWAS:
-
Genome-wide association studies
- IVW:
-
Inverse-variance weighted
- 25(OH)D:
-
25-Hydroxyvitamin D
- IV:
-
Instrumental variable
- SNP:
-
Single nucleotide polymorphism
- RCT:
-
Randomized controlled trial
- LD:
-
Linkage disequilibrium
- EAF:
-
Effect allele frequency
- CI:
-
Confidence interval
- MR-PRESSO:
-
MR-Pleiotropy Residual Sum and Outlier methods
- OR:
-
Odds ratio
- SMD:
-
Standardized mean difference
- CNS:
-
Central nervous system
References
Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, et al. 2016 American College of Rheumatology/European League against Rheumatism Classification Criteria for Primary Sjögren’s Syndrome: a consensus and data-driven methodology involving three international patient cohorts. Arthritis Rheumatol. 2017;69(1):35–45. https://doi.org/10.1002/art.39859.
Thorne I, Sutcliffe N. Sjögren’s syndrome. Br J Hosp Med (Lond). 2017;78(8):438–42. https://doi.org/10.12968/hmed.2017.78.8.438.
Bjordal O, Norheim KB, Rødahl E, Jonsson R, Omdal R. Primary Sjögren’s syndrome and the eye. Surv Ophthalmol. 2020;65(2):119–32. https://doi.org/10.1016/j.survophthal.2019.10.004.
Delli K, Dijkstra PU, Stel AJ, Bootsma H, Vissink A, Spijkervet FK. Diagnostic properties of ultrasound of major salivary glands in Sjögren’s syndrome: a meta-analysis. Oral Dis. 2015;21(6):792–800. https://doi.org/10.1111/odi.12349.
Negrini S, Emmi G, Greco M, Borro M, Sardanelli F, Murdaca G, et al. Sjögren’s syndrome: a systemic autoimmune disease. Clin Exp Med. 2022;22(1):9–25. https://doi.org/10.1007/s10238-021-00728-6.
Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjögren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61(6):554–8. https://doi.org/10.1136/ard.61.6.554.
Ramos-Casals M, Brito-Zerón P, Solans R, Camps MT, Casanovas A, Sopeña B, et al. Systemic involvement in primary Sjogren’s syndrome evaluated by the Eular-SS disease activity index: analysis of 921 Spanish patients (GEAS-SS Registry). Rheumatology (Oxford). 2014;53(2):321–31. https://doi.org/10.1093/rheumatology/ket349.
Callaghan R, Prabu A, Allan RB, Clarke AE, Sutcliffe N, Pierre YS, et al. Direct healthcare costs and predictors of costs in patients with primary Sjogren’s syndrome. Rheumatology (Oxford). 2007;46(1):105–11. https://doi.org/10.1093/rheumatology/kel155.
Rosen CJ. Clinical Practice. Vitamin D insufficiency. N Engl J Med. 2011;364(3):248–54. https://doi.org/10.1056/NEJMcp1009570.
Holick MF. Vitamin D Deficiency. N Engl J Med. 2007;357(3):266–81. https://doi.org/10.1056/NEJMra070553.
Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13(1):25–36. https://doi.org/10.1038/nrneurol.2016.187.
Erten Ş, Şahin A, Altunoğlu A, Gemcioğlu E, Koca C. Comparison of plasma vitamin D levels in patients with Sjögren’s syndrome and healthy subjects. Int J Rheum Dis. 2015;18(1):70–5. https://doi.org/10.1111/1756-185X.12298.
Agmon-Levin N, Kivity S, Tzioufas AG, López Hoyos M, Rozman B, Efes I, et al. Low levels of vitamin-D are associated with neuropathy and lymphoma among patients with Sjögren’s syndrome. J Autoimmun. 2012;39(3):234–9. https://doi.org/10.1016/j.jaut.2012.05.018.
Kuo CY, Huang YC, Lin KJ, Tsai TY. Vitamin D deficiency is associated with severity of dry eye symptoms and primary Sjögren’s syndrome: a systematic review and meta-analysis. J Nutr Sci Vitaminol (Tokyo). 2020;66(4):386–8. https://doi.org/10.3177/jnsv.66.386.
Smith GD, Ebrahim S. ‘Mendelian Randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. https://doi.org/10.1093/ije/dyg070.
Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33(1):30–42. https://doi.org/10.1093/ije/dyh132.
Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318(19):1925–6. https://doi.org/10.1001/jama.2017.17219.
Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep. 2017;4(4):330–45. https://doi.org/10.1007/s40471-017-0128-6.
Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the Strobe-MR statement. JAMA. 2021;326(16):1614–21. https://doi.org/10.1001/jama.2021.18236.
Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, EPIC- InterAct Consortium. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–52. https://doi.org/10.1007/s10654-015-0011-z.
Davey Smith G, Holmes MV, Davies NM, Ebrahim S. Mendel’s laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol. 2020;35(2):99–111. https://doi.org/10.1007/s10654-020-00622-7.
Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27(11):3253–65. https://doi.org/10.1681/asn.2016010098.
Collins R. What makes UK Biobank special? Lancet. 2012;379(9822):1173–4. https://doi.org/10.1016/s0140-6736(12)60404-8.
Revez JA, Lin T, Qiao Z, Xue A, Holtz Y, Zhu Z, et al. Genome-wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat Commun. 2020;11(1):1647. https://doi.org/10.1038/s41467-020-15421-7.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, et al. Finngen: unique genetic insights from combining isolated population and national health register data. medRxiv (2022):2022.03.03.22271360. https://doi.org/10.1101/2022.03.03.22271360.
Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. Phenoscanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851–3. https://doi.org/10.1093/bioinformatics/btz469.
Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64. https://doi.org/10.1093/ije/dyr036.
Papadimitriou N, Dimou N, Tsilidis KK, Banbury B, Martin RM, Lewis SJ, et al. Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat Commun. 2020;11(1):597. https://doi.org/10.1038/s41467-020-14389-8.
Staiger D, Stock JH. Instrumental variables regression with weak instruments. Econometrica. 1997;65(3):557–86. https://doi.org/10.2307/2171753.
Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–52. https://doi.org/10.1093/ije/dyq151.
Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–501. https://doi.org/10.1093/ije/dyt179.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. https://doi.org/10.1093/ije/dyv080.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14. https://doi.org/10.1002/gepi.21965.
Bowden J, Hemani G, Davey SG. Invited commentary: Detecting individual and global horizontal pleiotropy in Mendelian randomization-a job for the humble heterogeneity statistic? Am J Epidemiol. 2018;187(12):2681–5. https://doi.org/10.1093/aje/kwy185.
Ong JS, MacGregor S. Implementing MR-PRESSO and GCTA-GSMR for pleiotropy assessment in Mendelian randomization studies from a practitioner’s perspective. Genet Epidemiol. 2019;43(6):609–16. https://doi.org/10.1002/gepi.22207.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife (2018) 7. https://doi.org/10.7554/eLife.34408.
Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. https://doi.org/10.1038/s41588-018-0099-7.
de Lusignan S, Alexander H, Broderick C, Dennis J, McGovern A, Feeney C, et al. Atopic dermatitis and risk of autoimmune conditions: population-based cohort study. J Allergy Clin Immunol. 2022;150(3):709–13. https://doi.org/10.1016/j.jaci.2022.03.030.
Mavragani CP, Moutsopoulos HM. Sjögren’s syndrome: old and new therapeutic targets. J Autoimmun. 2020;110:102364. https://doi.org/10.1016/j.jaut.2019.102364.
Godala M, Sewerynek E, Maślach D, Krzyżak M, Gaszyńska E. Resting metabolic rate in women with endocrine and osteoporotic disorders in relation to nutritional status, diet and 25(OH)D concentration. Int J Environ Res Public Health (2022) 19(5). https://doi.org/10.3390/ijerph19053118.
Szodoray P, Horvath IF, Papp G, Barath S, Gyimesi E, Csathy L, et al. The immunoregulatory role of vitamins A, D and E in patients with primary Sjogren’s syndrome. Rheumatology (Oxford). 2010;49(2):211–7. https://doi.org/10.1093/rheumatology/kep374.
Hirsch S, Skripuletz T, Seeliger T, Witte T, Thiele T. Selenium deficiency is associated with polyneuropathy in primary Sjögren’s syndrome. Clinical Nutrition ESPEN. 2022;50:212–7. https://doi.org/10.1016/j.clnesp.2022.05.015.
Yin K, Agrawal DK. Vitamin D and inflammatory diseases. J Inflamm Res. 2014;7:69–87. https://doi.org/10.2147/jir.S63898.
Guzman-Prado Y, Samson O, Segal JP, Limdi JK, Hayee B. Vitamin D therapy in adults with inflammatory bowel disease: a systematic review and meta-analysis. Inflamm Bowel Dis. 2020;26(12):1819–30. https://doi.org/10.1093/ibd/izaa087.
Jiang J, Shao M, Wu X. Vitamin D and risk of ankylosing spondylitis: a two-sample Mendelian randomization study. Hum Immunol. 2022;83(1):81–5. https://doi.org/10.1016/j.humimm.2021.09.003.
Shoenfeld Y, Giacomelli R, Azrielant S, Berardicurti O, Reynolds JA, Bruce IN. Vitamin D and systemic lupus erythematosus - the hype and the hope. Autoimmun Rev. 2018;17(1):19–23. https://doi.org/10.1016/j.autrev.2017.11.004.
Jiang X, Alfredsson L. Modifiable environmental exposure and risk of rheumatoid arthritis-current evidence from genetic studies. Arthritis Res Ther. 2020;22(1):154. https://doi.org/10.1186/s13075-020-02253-5.
Schulz KF, Grimes DA. Case-control studies: research in reverse. Lancet. 2002;359(9304):431–4. https://doi.org/10.1016/s0140-6736(02)07605-5.
Lawlor DA, Davey Smith G, Kundu D, Bruckdorfer KR, Ebrahim S. Those confounded vitamins: what can we learn from the differences between observational versus randomised trial evidence? Lancet. 2004;363(9422):1724–7. https://doi.org/10.1016/s0140-6736(04)16260-0.
Acknowledgements
Our thanks go out to the original GWAS participants and investigators, as well as to the FinnGen research group, and the related consortiums for sharing and managing the summary statistics.
Patient and public involvement
Patients and the public were not involved in the design, conduct, reporting, and dissemination plans of this manuscript.
Funding
This research received no funding.
Author information
Authors and Affiliations
Contributions
MZ, FRW, and MPW conceived and designed the study. YG, YYL, ZMW, SW, and GQF collected the data. MZ and HL analyzed the data and results. MZ and HL completed the writing of the manuscript. MZ, FRW, and MPW reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Data were collected from public databases. And there is no ethical approval necessary.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Meng Zhao, Feiran Wei these authors contributed equally to this work and share the first authorship.
Supplementary Information
Additional file 1:
Supplementary Table 1. Summary of genetic variants (n=81) used to estimate the effect of serum vitamin D on SS in MR analyses. Supplementary Table 2. Summary of genetic variants (n = 4) used to estimate the effect of SS on serum vitamin D in MR analyses.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, M., Wei, F., Li, H. et al. Serum vitamin D levels and Sjogren’s syndrome: bi-directional Mendelian randomization analysis. Arthritis Res Ther 25, 79 (2023). https://doi.org/10.1186/s13075-023-03062-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13075-023-03062-2