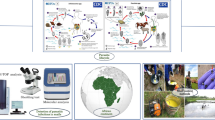
Abstract
Background
Snail-borne trematodes afflict humans, livestock, and wildlife. Recognizing their zoonotic potential and possible hybridization, a One Health approach is essential for effective control. Given the dearth of knowledge on African trematodes, this study aimed to map snail and trematode diversity, focusing on (i) characterizing gastropod snail species and their trematode parasites, (ii) determining infection rates of snail species as intermediate hosts for medically, veterinary, and ecologically significant trematodes, and (iii) comparing their diversity across endemic regions.
Methods
A cross-sectional study conducted in 2021 in Chiredzi and Wedza districts in Zimbabwe, known for high human schistosomiasis prevalence, involved malacological surveys at 56 sites. Trematode infections in snails were detected through shedding experiments and multiplex rapid diagnostic polymerase chain reactions (RD-PCRs). Morphological and molecular analyses were employed to identify snail and trematode species.
Results
Among 3209 collected snail specimens, 11 species were identified, including schistosome and fasciolid competent snail species. We report for the first time the invasive exotic snail Tarebia granifera in Zimbabwe, which was highly abundant, mainly in Chiredzi, occurring at 29 out of 35 sites. Shedding experiments on 1303 snails revealed a 2.24% infection rate, with 15 trematode species identified through molecular genotyping. Five species were exclusive to Chiredzi: Bolbophorus sp., Schistosoma mansoni, Schistosoma mattheei, Calicophoron sp., and Uvulifer sp. Eight were exclusive to Wedza, including Trichobilharzia sp., Stephanoprora amurensis, Spirorchid sp., and Echinostoma sp. as well as an unidentified species of the Plagiorchioidea superfamily. One species, Tylodelphys mashonensis, was common to both regions. The RD-PCR screening of 976 non-shedding snails indicated a 35.7% trematode infection rate, including the presence of schistosomes (1.1%) Fasciola nyanzae (0.6%). In Chiredzi, Radix natalensis had the highest trematode infection prevalence (33.3%), while in Wedza, R. natalensis (55.4%) and Bulinus tropicus (53.2%) had the highest infection prevalence.
Conclusions
Our xenomonitoring approach unveiled 15 trematode species, including nine new records in Zimbabwe. Schistosoma mansoni persists in the study region despite six mass deworming rounds. The high snail and parasite diversity, including the presence of exotic snail species that can impact endemic species and biomedically important trematodes, underscores the need for increased monitoring.
Graphical Abstract

Similar content being viewed by others
Background
Millions of people in approximately 90 countries suffer from snail-borne trematodiases [1, 2]. For example, schistosomiasis, a widespread and neglected tropical disease caused by species of the genus Schistosoma, holds significant prevalence, morbidity, and socioeconomic implications [2]. Moreover, trematodiases like schistosomiasis, fasciolosis, echinostomiasis, and amphistomosis also form an economic burden on the livestock industry as they lead to production losses and mortality in livestock [3]. For example, fasciolosis alone causes global economic losses of over US$3.2 billion per year [4]. Although the impact of trematodes on wildlife populations is poorly understood, high parasite load may lead to mortality which threatens conservation efforts [5, 6].
Some trematodiases are regarded as zoonotic. Fasciolosis is primarily of concern in domestic and wild animals, but human cases have been reported in 42 countries, mostly in Asia and South America [7,8,9]. Fasciola hybrids commonly occur and are capable of infecting humans [10, 11]. Hybridization between human and animal schistosome species has also been reported [12,13,14], demonstrating the importance of animal reservoirs in disease ecology. Therefore, it is recommended to study trematodiases within a One Health framework, where the health of humans, livestock, and wildlife is considered intrinsically linked and equally important for mapping, detecting high-risk locations, and improving disease control [13, 15,16,17].
In Zimbabwe, trematodiases due to schistosomes, fasciolids, and amphistomes have been reported [18,19,20,21,22]. The presence of human schistosomiasis in Zimbabwe has long been documented [23,24,25] and the latest national survey revealed the disease is present in at least 91% of all districts, being most prevalent in Chiredzi, Wedza, and Shamva districts [24]. Human fasciolosis has also been described in Zimbabwe [26], but no recent cases have been reported. Records of animal fasciolosis due to Fasciola gigantica in livestock [27], Fasciola hepatica in antelope (Hippotragus niger) and a duiker (Sylvicapra grimmia) [19] and recently Fasciola nyanzae in the common hippopotamus [28] have been published.
Recent malacological surveys in the country revealed a high diversity of trematodes [28,29,30,31], many of which could not be identified through molecular barcoding techniques because of the gross underrepresentation of trematode reference sequences in public databases, the so-called “barcoding void” [30]. Failure to identify trematodes hinders prompt interventions, further reinforcing their neglected status.
For this study, the Chiredzi and Wedza districts in Zimbabwe were chosen because of reported high schistosomiasis prevalence in 2014 [24]. Revisited after a decade, we extended our study to all trematode species and their associated freshwater snail species, aligning with the One Health approach. This study sought to (i) characterize gastropod snail and trematode diversity and (ii) determine infection rates in snails acting as intermediate hosts for medically, veterinary, and ecologically significant trematodes.
Methods
Study area
The cross-sectional malacological survey was conducted in August 2021, during which 35 sites in Chiredzi and 21 in Wedza were sampled. Chiredzi, in southeastern Zimbabwe, is renowned for sugarcane farming, notably around the Chiredzi and Runde rivers, with major plantations at Hippo Valley Estates, Triangle, and Mkwasine. The region's irrigation relies on canal-facilitated flooding. With mean annual rainfall of 582 mm, Chiredzi is semi-arid, experiencing a hot-wet season (mid-November to April), cool-dry season (May to late July), and hot-dry season (August to mid-November). Daily temperatures frequently surpass 32 °C, reaching peaks of at least 45 °C, making it a water-stressed region [32]. Over 70% of Chiredzi is dedicated to wildlife conservation, with Gonarezhou National Park (GNP), Malilangwe Wildlife Reserve (MWR), and Save Conservancy as principal custodians.
Wedza, on the other hand, has a temperate highland tropical climate that typically receives about 675 mm of precipitation spread across 143 rainy days annually [32]. The annual average temperature is 21.5 °C. The district is bound by the Save River in the west and the Ruzave River in the east. Other rivers include Chineyi, Mhare, Nyamidzi, and Nyamhembe.
In Chiredzi, we focused on the protected area of MWR and human-impacted regions, including Chipimbi, Hippo Valley, and Triangle. In Wedza district, the primary focus was on the Imire Rhino and Wildlife Conservation area (referred to as Imire), dedicated to wildlife preservation and partially to tobacco farming. Selection of sampling sites in both regions was based on features indicative of potential transmission areas, including areas with frequent human and/or animal water contact. Refer to Additional file 1: Table S1 for detailed site characteristics.
Field collection of snails and trematodes
Sampling occurred in a standardized way across all sites, by scooping for 30 min by two persons per site with a scoop net of 1-mm mesh size with a 2-m-long handle. GPS coordinates were recorded at each sampling point. Live snails were transported in ambient water to the field laboratory for sorting, counting, and identification to species or genus level using keys by Mandahl-Barth [33], Brown [34], and Appleton & Miranda [35]. Snails were put in 24-well tissue culture plates with rested borehole water and kept in the dark overnight as per Gumble et al. [36]. The morning after, the snails were exposed to light for several hours and visually checked for cercariae. Emerging cercariae were individually isolated on microscopic slides and photographed and measured using BMS camera (model number: XFCAM1080PHD) and associated BMS pix3 software (version 3.7.8942.20170412) with attention on morphological traits (i.e. oral and ventral suckers, stylet, eyespots, collar spines, tail, and finfold) to classify them per morphotype using keys by Frandsen & Christensen [37]. Shedding snails and cercariae were individually stored, while non-shedding snails were pooled by species per site in 80% ethanol in larger 50-ml ethanol tubes for transportation to Belgium for molecular analysis.
Snail and trematode DNA extraction
A sterile needle was used to make an incision in the shell apex and subsequently push the soft tissue out of the shell. Next, the isolated tissue was dried using absorbent paper. DNA was extracted using the E.Z.N.A.® Mollusc DNA Kit (OMEGA Bio-Tek, Norcross, GA, USA) according to the manufacturer’s protocol and eluted through two elution steps of 50 μl, totaling 100 μl of DNA extract. Only medical and veterinary important snails including planorbids and lymnaeids were subjected to DNA extraction as they would later be examined for infection using PCR (see next section). For each of the remaining snail species, one specimen per species was extracted for phylogenetic analyses.
The DNA of individual cercariae was extracted using a proteinase K-based lysis buffer as described in Ziętara et al. [38]. In short, 10 μl of the prepared proteinase K lysis buffer was added to a single cercaria in 10 μl of MilliQ water (Merck, Darmstadt, Germany). Next, a two-step incubation followed at 65 °C for 25 min and at 95 °C for 10 min after which DNA extracts were stored undiluted at − 20 °C.
Molecular infection diagnosis in snails
Up to 30 specimens of biomedically important snails (i.e. belonging to the Bulinus and Biomphalaria genera or the family of the Lymnaeidae) were subsampled per species, per site for DNA extraction and tested for infection through the multiplex rapid diagnostic polymerase chain reaction (RD-PCR) methods described by Schols et al. [39] and Carolus et al. [29]. These approaches enable the detection of (pre)patent infections of any trematode species, including Schistosoma and Fasciola species, in snail DNA extracts. Briefly, both methods are based on the amplification of multiple markers of different lengths in a single PCR reaction: an internal control that amplifies snail 18S rDNA to confirm DNA extraction and PCR success, a general trematode primer pair designed to amplify an 18S rDNA fragment of all trematode genera that have a reference sequence available in GenBank, and primers for Schistosoma or Fasciola- specific amplification based on the internal transcribed spacer 2 (ITS2) and nuclear-repeat region respectively. All used primers and PCR protocols are described elsewhere [29, 39]. Samples that were positive for Schistosoma spp. infection were subsequently analyzed using a species-specific multiplex PCR according to the two-step approach described by Schols et al. [39]. This multiplex PCR differentiates between several schistosomes of veterinary and medical importance (i.e. Schistosoma haematobium, S. mansoni, S. mattheei, and S. bovis/S. curassoni/S. guineensis) by generating species-specific COI amplicons of different lengths.
PCR and DNA sequencing of snails and trematodes
Snails were identified using the COI marker whereas trematodes were identified by targeting the COI, 18S rDNA, and the ribosomal internal transcribed spacer (ITS) 1 and 2 markers. The primers and resulting amplicon lengths are listed in Table 1, together with those for snail and trematode identification. PCR assays were run in a 20 μl reaction volume using the Qiagen™ Taq DNA polymerase kit comprising 0.6 mM dNTP mix, 2 µl 10 × PCR buffer, 1.5 mM MgCl2, 0.12 µl Taq Polymerase at 5 units/µl, 0.8 μM of each primer, and 2 μl DNA extract (1:10 diluted). The PCR cycling parameters varied depending on the respective primers used following Schols et al. [28], and the PCR amplifications were performed in a Biometra® Tprofessional Thermal Cycler. PCR products were visualized by gel electrophoresis on a 2% agarose gel containing Midori Green Direct® staining using a UV transilluminator. Samples exhibiting a clear and bright PCR product were purified using the ExoSAP (Fermentas™) PCR purification protocol and sequenced using the BigDye® chemistry by Macrogen™.
Phylogenetic analysis
Sequences were processed in Geneious Prime® (version 2023.1.1), carefully addressing ambiguities by examining complete chromatograms before and after consensus generation to ensure optimal sequence quality. The Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/) was used to probe species identity for all sequences of sufficient quality. Sequences with ambiguous species identity (pairwise distance > 5%, and various species in top BLAST results) were further analyzed through phylogenetic reconstructions with sequences from closely related taxa. Those sequences were mined from GenBank and aligned with MAFFT v.7 implemented online (https://mafft.cbrc.jp/alignment/server; [40]) with default settings. As an extra step, the least stringent settings in Gblocks available online at NGPhylogeny.fr (https://ngphylogeny.fr/; [41]) were applied to all ITS and 18S rDNA alignments. The model selection analysis was run in MEGA (version 11.0.13) to evaluate the most suitable substitution model based on the Akaike information criterion (AIC) and Bayesian information criterion (BIC) values from the model list. The suitable substitution models for all alignments and phylogenetic analyses performed in this study are described in the captions of their respective tree figures and supplementary files.
Lastly, the IQ-tree web servers (http://iqtree.cibiv.univie.ac.at) were used to calculate a maximum likelihood (ML) tree, and nodal support was assessed by performing 10,000 bootstrap replicates [42]. The resulting trees were rooted with the respective outgroups in Figtree (version 1.4.4) and exported to TreeGraph 2 (version 2.15.0–887 beta) [43] where nodes with support values < 70% were collapsed. The final tree was edited in Inkscape (version 1.3) where ill-placed nodal support values were adjusted, and species names were italicized.
Results
Observed freshwater snail species
Eleven different species of freshwater snails were observed from both Chiredzi and Wedza: Gyraulus sp., Melanoides tuberculata, Tarebia granifera, Biomphalaria pfeifferi, Pseudosuccinea columella, Physella acuta, Radix natalensis, Bulinus globosus, B. truncatus, B. forskalii, and B. tropicus (Fig. 1). The COI-based BLAST results from all our freshwater snails were satisfactorily robust for species identification: BLAST identity ≥ 99% with query cover of 100%, sequence lengths of + 300 bp, and HQ ≥ 95%. Our 467-bp B. pfeifferi sequences from both Chiredzi and Wedza had 100% nucleotide similarity with Malawian specimens (“GenBank: OR880274”). Similarly, a 100% match was observed between our 483 bp T. granifera sequences and those reported from China (“GenBank: MZ662113”) and Thailand (“GenBank: MK000375”). The sequences of our R. natalensis (467 bp) from Wedza matched perfectly with previous Zimbabwean records at least 150 km up north in Mazowe Dam (Mazowe district) and Mwenje Dam (Chiweshe district) (“GenBank: MT992958”). Our P. columella sequences (483 bp) indicated 100% similarity with records of the invasive haplotype across Africa (South Africa: “GenBank: MN601428” and Egypt: “GenBank: LC015522”), Australia (“GenBank: AY227366”), Europe (Spain: “GenBank: KP242798”), and North America (New Mexico, USA: “GenBank: NC_042905”). Similarly, our P. acuta sequence of 314 bp was identical to submissions from Asia (Japan: “GenBank: LC429395”), the Middle East (Iran: “GenBank: KT280442”), and the Americas (New York, USA: “GenBank: MN164018”). Our M. tuberculata sequence was of low quality (HQ 88.6%); hence, only morphology was used. The Planorbidae reference sequences from GenBank overlapped at least 378 bp with our partially sequenced COI marker. Using this fragment in phylogenetic inference, the species status and evolutionary relationships of B. globosus, B. truncatus, B. forskalii, and B. tropicus specimens collected in Chiredzi and Wedza were confirmed (Fig. 2). The smallest COI pairwise distances between our Gyraulus sp. (439 bp) and the reference sequences from GenBank were 3.2% (G. costulatus from Senegal “GenBank: OP811018”) and 4% (G. connollyi from South Africa “GenBank: KC495776”) (results not shown). However, our specimen clustered with G. costulatus (from Senegal “GenBank: OP811018”) in the phylogenetic tree (Fig. 2).
All 11 snail species sampled in Chiredzi and Wedza districts of Zimbabwe. Photographs were captured using a Canon EOS 600D camera with a Macro Photo Lens to capture images of specimens that were fixed on a dark clay platform presented in apertural, lateral, and sometimes apical view. The images were stacked using Zerene Stacker™ software and processed in Adobe Photoshop®, removing the background and combining the front and rear perspectives into one picture with a consistent scale size. A Bulinus tropicus; B B. globosus; C B. truncatus; D B. forskalii; E Biomphalaria pfeifferi; F Gyraulus sp.; G Radix natalensis; H Pseudosuccinea columella; I Physella acuta; J Melanoides tuberculata; K Tarebia granifera. The white line beneath the photo of each snail represents a scale bar of 5 mm
Maximum likelihood phylogenetic trees of the family Planorbidae using COI (655 bp) sequences and using the General Time Reversible (GTR) model [95] with discrete gamma distribution ([+ G] = 0.24). Bootstrap values (10,000 replicates) > 70 are shown next to the branches. GenBank sequences are displayed with their accession number (not italicized). Sequences without accession number were obtained during this study and can be linked to pictures of freshwater snails shown in the Fig. 1. The number or letter after the S-prefix denotes site name and respective the code name is given (BPF = B. pfeifferi, FOR = B. forskalii, BGL = B. globosus, TRU = B. truncatus, TRO = B. tropicus, and GYR = Gyraulus sp.)
Abundance and spatial distribution of freshwater snails
A total of 3209 snails were collected in Chiredzi (n = 1883) and Wedza (n = 1326). This record, however, excludes T. granifera, which was by far the most abundant snail species in Chiredzi with counts often exceeding 2500 individuals per site. Tarebia granifera also had the highest prevalence, occurring at 29 out of 35 sampled sites. Moreover, it seems to be most abundant and prevalent in areas with proximity to or run-off from irrigated sugarcane farms. As illustrated in Fig. 3, T. granifera is most abundant in sites that drain from Hippo Valley Estates sugarcane fields (e.g., sites 16–18 and 31–44), as well as site 6 (Chipimbi River) and 14 (Chiredzi River). However, sites 9–13 are located in Hippo Valley Estates but are not dominated by T. granifera. In Chiredzi we found nearly all 11 freshwater snail species mentioned above, except for Gyraulus sp. and B. tropicus. Counts for medically and veterinary important snails in Chiredzi are as follows: B. globosus (n = 305), B. truncatus (n = 259), B. forskalii (n = 2), B. pfeifferi (n = 118), P. columella (n = 40), and R. natalensis (n = 23). In addition to T. granifera, other invasive species found in Chiredzi included P. acuta (n = 858) and M. tuberculata (n = 278). In Wedza, all species mentioned above were observed except for T. granifera and B. forskalii. Counts for medically and veterinary important snails in Wedza are as follows: B. globosus (n = 55), B. truncatus (n = 20), B. tropicus (n = 466), B. pfeifferi (n = 86), P. columella (n = 6), and R. natalensis (n = 401). For other species we found P. acuta (n = 288), Gyraulus sp. (n = 1), and M. tuberculata (n = 3). See Additional file 1: Table S2 for the detailed number of snail species and snail specimens per site. Here, the data also show that sites with high T. granifera abundance harbor relatively low numbers of schistosome-competent snails.
Distribution of snails collected during this study. A The sampling map for Chiredzi district including wetlands in the Malilangwe and juxtaposed Hippo Valley and Triangle sugarcane plantations. Most sites have a low diversity of only three or fewer species. Of special note is the widespread coverage by the invasive Tarebia granifera (represented by a navy blue color) including sites 6, 14, and 16 to 18 as well as 31 to 44 (pie charts shown in red box as sites are too close to each other). B Sampling map of Wedza (Imire). Radix natalensis (brown color) was quite widespread and found in almost every site except the three most southerly sites mostly dominated by bulinids (Bulinus globosus, B. tropicus, and B. truncatus). Also widely distributed is Physella acuta (green color). Sampling sites in green circle are too close together and are projected on the right-hand side for map. See Additional file 1: Table S2 for the detailed number of snails per site. The bottom left side shows the position of Imire (Wedza) and Chiredzi on the Zimbabwean map
Identified trematode species released from freshwater snails
In total, 1743 snail specimens belonging to the following seven species—B. pfeifferi, B. truncatus, B. forskalii, B. globosus, B. tropicus, P. columella, and R. natalensis—were tested for shedding. Eleven cercarial morphotypes were recovered. Four were exclusive to Chiredzi: ophthalmocercariae, longifurcate-pharyngeate distome cercariae (subtype I), amphistome type cercariae, and brevifurcate-apharyngeate distome (subtype I) coded as Morph I to Morph IV, respectively (Fig. 4). Longifurcate-pharyngeate distome cercariae (subtype II) (Morph V) was the only overlapping morphotype between Chiredzi and Wedza. The remaining six morphotypes were only exclusive to Wedza: plagiorchiid (subtype I and II), strigea type cercaria, echinostome type cercaria, longifurcate-pharyngeate distome cercaria (subtype III), and brevifurcate-apharyngeate monostome cercaria (subtype III) coded as Morph VI to Morph XI, respectively (Fig. 4).
Cercariae identification from snail species in Chiredzi and Wedza. A Photographs depict 11 distinct cercarial morphotypes classified based on morphological characteristics following Frandsen and Christensen [37]. Each morphotype is represented by one specimen, with Morph I-IV from Chiredzi, Morph VI-XI from Wedza, and Morph V found in both regions. B Molecular genotyping refined the identification, ultimately splitting Morph IV, V, and IX into two, resulting in 14 unique species. C Molecular markers (COI, 18S rDNA, and ITS) were employed for cercariae identification, with successful markers denoted by a tick symbol, while unsuccessful ones are marked with an ‘x’
For every snail that released cercariae, sequencing was performed on one cercarial specimen per morphotype (Additional file 1: Table S3), and only then were we able to assign sub-morphotypes for ease of interpretation of results. Three markers (COI, 18S, and ITS) were used for the sequence-based analysis where the highest BLAST results and phylogenetics were inferred (Additional file 1: Table S3). At least 14 different trematode species were detected from the 11 cercarial morphotypes. Four morphotypes were identified to species level, i.e. Schistosoma mattheei (Morph IV), S. mansoni (Morph IV), Tylodelphys mashonensis (Morph V), and Stephanoprora amurensis (Morph IX). Six morphotypes were identified to genus level, i.e. Bolbophorus sp. (Morph II), Calicophoron sp. (Morph III), Uvulifer sp. (Morph V), Trichobilharzia sp. (Morph VIII), Echinostomata sp. (Morph IX), and Spirorchid sp. (Morph XII). Four species were identified to (super) family level, i.e. Strigeidae (Morph X) and Plagiorchioidea (Morph I, Morph VI, and Morph VII). Figure 4 shows the markers used to determine the taxa identity but see Additional file 1: Table S3 which compiled exact methods used including phylogenetic analysis in Fig. 5 and Additional file 1: Fig. S5).
Maximum likelihood phylogenetic tree of superfamilies Diplostomoidea, Echinostomatoidea and Plagiorchioidea using COI (655 bp) and using the General Time Reversible (GTR) model [95] with discrete gamma distribution ([+ G] = 0.59) and invariant sites ([+ I] = 0.34). Nodal support is indicated as bootstrap percentages (10,000 bootstraps). GenBank sequences are displayed with their accession number. Sequences without accession number (labeled as Morphs) were obtained during this study and can be linked to pictures of released larval trematodes (cercariae) shown in Fig. 4. While an attempt was made to identify nine cercarial morphotypes, only two were identified with 100% match due to the barcoding void
Trematode species identified through RD-PCR
In Chiredzi, overall, nine out of 726 snails (1.24%) were shedding. Of these shedding snails, we identified five cercarial morphotypes later differentiated into six different species through molecular genotyping (see above). Next, we used RD-PCR to determine trematode infection in snails. Of the 360 snails from Chiredzi examined by RD-PCR, 44 were found to have a trematode infection, with prevalence in each snail species ranging from 3 to 33%. Four of these infections were Schistosoma spp. and two were Fasciola sp. infections. The highest trematode infection prevalence was found in R. natalensis at 33.3% (11 out of 33) (Table 2). In Wedza, 30 out of 1017 snails (2.95%) were shedding. Of these shedding snails, we identified seven cercarial morphotypes later differentiated into nine different species through molecular genotyping (see above). Of the 616 snails from Chiredzi examined by RD-PCR, 306 were found to have a trematode infection, with prevalence per snail species ranging from 25 to 55%. None of the infections could be attributed to either a Schistosoma or Fasciola sp. infection. The highest infection rate was detected in R. natalensis at 55.4% (159 out of 287), followed by B. tropicus at 53.2% (107 out of 201) (Table 2).
For those snail specimens with a positive schistosome signal in the RD-PCR, a second, species-specific multiplex PCR test was conducted to discriminate among Schistosoma species. Schistosoma mansoni was only found at site 6 (Chipimbi River, Chiredzi) with an infection prevalence of 11.1% (2/18) in B. pfeifferi. Schistosoma mattheei was found in B. globosus at site 6 (Chipimbi River) and site 20 (Masvisvidza Dam) with a prevalence of 9.1% (1/11) and 12.5% (1/8), respectively. No Schistosoma haematobium infection was found in this study. Trematode infections of two P. columella snails (not shedding) from site 19 (Gungwa River, Chiredzi) that appeared positive after RD-PCR (see below) were subjected to COI sequencing. The resultant trematode sequences (399 bp) were 100% identical to Fasciola nyanzae from South Africa (“GenBank: ON661099”) and 99.8% to F. nyanzae from Zimbabwe (“GenBank: MT909542”).
Discussion
We studied freshwater snail species and their trematode parasites in a region highly endemic for schistosomiasis. A high snail species diversity including new records, and a plethora of trematode species were found.
Freshwater snail diversity and spatial distribution
Our study presents a comprehensive malacological survey conducted in two significant biodiversity conservation areas and their adjacent human-influenced zones in Zimbabwe. We identified 11 species of freshwater snails in Chiredzi and Wedza. The snail species diversity in our study aligns with local findings by Schols et al. [30] and Muzarabani et al. [31], reporting 12 and seven species, respectively, in Zimbabwe. Comparable diversity has been reported in other regional studies: eight species in KwaZulu-Natal province, South Africa [44], five species in southwest Ethiopia [45], and eight species in north Cameroon [46]. Furthermore, studies in Denmark recovered 10 species [47], while 15 species were reported from North Rhine-Westphalia, Germany [48].
The identified snail species are Gyraulus sp., Melanoides tuberculata, Tarebia granifera, Biomphalaria pfeifferi, Pseudosuccinea columella, Physella acuta, Radix natalensis, Bulinus globosus, Bulinus truncatus, Bulinus forskalii, and Bulinus tropicus. Four of these species, B. pfeifferi, B. globosus, B. truncatus, and B. forskalii, hold medical significance as intermediate hosts for urinary and intestinal schistosomiasis. The bulinid species, including B. tropicus, can also transmit bovine schistosomes [49] and stomach flukes [31]. Other snail species, including R. natalensis and P. columella, act as intermediate hosts for liver flukes [50]. Four species–T. granifera, M. tuberculata, P. columella, and P. acuta–are alien to Zimbabwe (see below).
Tarebia granifera, P. acuta, and R. natalensis emerged as the most widespread species, with T. granifera being the most abundant. Gyraulus sp. and B. forskalii were the rarest and least abundant species. All except one of these species have very recently been documented in previous records for Zimbabwe [28,29,30,31], with T. granifera being a new record.
Bulinus forskalii exclusively inhabited Chiredzi, while B. tropicus and Gyraulus sp. were unique to Wedza, potentially influencing variations in parasite fauna between the two areas, as discussed in detail below. Found only in Chiredzi, the invasive snail T. granifera exhibited particularly high abundance, reaching up to 2500 specimens per site in areas with runoff from sugarcane cultivation. This aligns with existing documentation highlighting the highly eutrophic and fragmented nature of Chiredzi's drainage system [51]–a trait known to attract highly invasive snail species [34, 52]. Our results indeed show that anthropogenically disturbed sites appear more vulnerable to invasive species, in line with other findings [29, 34, 52]. In this case, run-off from sugarcane cultivation appears to facilitate T. granifera populations, while native species of medical importance like Biomphalaria and Bulinus snail species were absent in these sites. Generally, there is higher diversity (at least three species) in the conserved areas, specifically in Lake Malilangwe and in Imire, compared to the cultivated regions mentioned earlier. This observation aligns with literature indicating that human-disturbed ecosystems often witness a decline in macroinvertebrate abundance and diversity [53], although several studies emphasize that schistosome-competent snails thrive in these impacted habitats [54].
Exotic invasive snails: friend or foe?
Accounts for several exotic invasive snails in southern Africa have been published [29, 55,56,57]. Aquaculture, recreation, transport, and trade globalization may intentionally or accidentally bolster the transport of exotic invasive freshwater snails, of which some species can become invasive, thereby disrupting local ecosystems [58,59,60]. One such example is T. granifera, a strongly invasive snail species native to regions of Southeast Asia and Oceania [61]. The first report of T. granifera in Africa was in 1999 in northern KwaZulu-Natal, South Africa, followed by rapid colonization of the eastern part of the country and neighboring Eswatini [59, 60, 62]. To the best of our knowledge, this is the first report of T. granifera identified at species level in Zimbabwe and our sequencing results revealed that our haplotype was (nearly) identical with specimens from Malawi, French Polynesia, Alabama (USA), Thailand, and Timor. Unfortunately, at the present moment, no T. granifera COI sequences from South Africa could be found on GenBank making it difficult to trace the source of introduction. In this study, T. granifera covered, by the thousands per unit area, nearly all waterbodies in Chiredzi directly linked with the sugarcane plantations. It has been found that T. granifera can reach densities of up to 10,000 individuals/m2, displacing most endemic snail species, a ‘foe’ in aquatic biodiversity conservation, as was the case in wetlands in Puerto Rico [63], Venezuela [64], and South Africa [55, 65]. However, this potentially creates opportunities for biological control against schistosome/Fasciola-competent snail species, thereby becoming a ‘friend’ from a public health perspective [66, 67]. Competitive exclusion might, therefore, explain the absence of endemic snail species at sites where T. granifera is present, especially schistosome-competent snails, which were either scarce or entirely absent in the presence of T. granifera. Conversely, there was an increased occurrence of schistosome-competent snails in the absence or low numbers of T. granifera.
Having a North American origin, the invasive exotic P. columella was introduced in South Africa together with exotic aquatic crops in the 1960s [68, 69]. It was described for the first time in Zimbabwe three decades ago [34], and until then only recently reported in the country’s largest reservoir, Lake Kariba [29], as well as in Chiweshe [30]. The P. columella isolates from Chiredzi and Wedza were genetically identical to isolates from Lake Kariba (Zimbabwe), South Africa, Australia, Colombia, Egypt, Spain, and the USA, supportive of the ‘flash invasion’ scenario described for this species [69]. Pseudosuccinea columella has been reported as a highly compatible host for F. gigantica in South Africa [70] and Egypt [50] and for F. nyanzae in Zimbabwe [28]. Therefore, P. columella is a ‘foe’ associated with the increased transmission of endemic Fasciola species–representing a parasite spillback phenomenon whereby a non-indigenous snail host is infected by a native parasite [29]. Also, in this study some P. columella specimens were infected with F. nyanzae, further emphasizing the increased disease risk posed by this snail (see below).
Finally, P. acuta, the third exotic species encountered in this study, was also very widespread, both in Wedza and Chiredzi. Here, we found it in 26 of the 56 sites, with abundance ranging from 1 to 224. It was however never found to be infected. Originating from North America, P. acuta has achieved cosmopolitan status through its successful dispersion facilitated by the ornamental aquarium trade [71]. It has proliferated across various global regions, encompassing Africa, Asia, Australia, Europe, and South America [58, 71,72,73,74]. Documented instances indicate its displacement of populations of Bulinus and Biomphalaria species in specific African regions, such as Nigeria, Ghana, and Southern Mozambique [34, 58, 75]. This ecological replacement positions P. acuta as a potential ‘friend’ in mitigating schistosomiasis, prompting considerations for its integration into biological control strategies [75, 76]. However, given its invasive tendency, prudent management practices are imperative to prevent Physella populations from surpassing ecologically crucial thresholds and therefore becoming a detriment to biological diversity conservation as warned in Lawton et al. [71].
Biomedically important trematodes with One Health implications
Schistosomiasis and fascioliasis, two diseases of public health and veterinary importance, are of great concern both worldwide and in Zimbabwe. Biomphalaria pfeifferi is the primary intermediate host for S. mansoni that causes intestinal schistosomiasis in sub-Saharan Africa [77]. Additionally, its relatively high numbers across our sampled regions align with the high prevalence of intestinal schistosomiasis among children, with rates of 43.7% in Chiredzi and 32.3% in Wedza, as reported by Midzi et al. [24]. We found S. mansoni in the intermediate snail host Bi. pfeifferi in Chiredzi with an overall prevalence of 1.7%. Similarly, we also found S. mattheei, a cattle schistosome, infecting B. globosus with an overall prevalence of 0.7%. These prevalence levels are on the lower side, but a few infected snail specimens are sufficient for sustaining local transmission. For example, low prevalence of S. haematobium in Bulinus spp. (1.9%) was recorded in Shamva district, Zimbabwe [78], yet the region has a very high human schistosomiasis prevalence of > 50% [24]. This is contrary to the high overall infection rate of 11.1% of S. mattheei in Bulinus spp. found in Mazowe district, Zimbabwe [30]. However, the low occurrence could also be attributed to our cross-sectional sampling design, offering only a single snapshot in time. As such, longitudinal studies are needed to account for seasonal dynamics in disease transmission [79]. Although the present study did not find snails infected with S. haematobium, the high human prevalence between 10 and 49% in both regions as reported by Midzi et al. [24] and the local presence of B. globosus in both districts imply a substantial risk for urogenital schistosomiasis.
Through COI-based molecular barcoding, we found P. columella infected with F. nyanzae at a prevalence of 2.3%. Our record is, therefore, the third reported district in Zimbabwe after Chiweshe [30] and Kariba [28] with this parasite. Fasciola nyanzae is considered to be an exclusive parasite of the common hippopotamus [80, 81]. However, these recent reports partially overlap with livestock areas, raising the potential risk for hybridization with other fasciolids, a phenomenon frequently reported in this group [10, 11]. We also found Calicophoron sp. infecting one B. truncatus at one site in Chiredzi. This amphistome species of veterinary health concern was reported in Southern Africa, but until now its local intermediate snail host was not yet determined [21].
While no hybrids were discovered in our study, the medically significant trematodes we examined have been previously recognized for their zoonotic potential. These parasites possess the capability to utilize both human and animal reservoir systems, posing significant concerns for their study and control. For instance, certain species of wild rodents serve as reservoirs for S. mansoni in endemic areas in Africa and the Caribbean [82, 83]. Additionally, we encountered S. mattheei, a cattle parasite, reported to naturally infect free-ranging baboons in Zambia [84]. Therefore, it is imperative to maintain a robust One Health approach to comprehensively assess trematode diversity and hybridization in the sampled region, particularly due to the coexistence of humans and animals in infested areas. Indeed, these monitoring efforts can identify areas of human-animal trematode overlap, informing treatment strategies. For instance, according to Laidemitt et al. [85], treating cattle for the stomach fluke Calicophoron sukari could affect human intestinal schistosomiasis prevalence, as the stomach fluke larvae impede the development of S. mansoni in Biomphalaria snails.
Similar intermediate hosts miles apart but many different parasites?
Ultimately, our study yielded 15 trematode species as refined by molecular genotyping. Among these, six were previously reported in Zimbabwe: F. nyanzae [28], S. mattheei [30], S. mansoni [30], T. mashonensis [86], Calicophoron sp., and a Strigeidae species [87]. The remaining nine represent new species records for Zimbabwe: Bolbophorus sp., Trichobilharzia sp., Spirorchid sp., S. amurensis, Uvulifer sp., three Plagiorchioidea spp., and one Echinostomatoidea sp. None of these parasites have been reported in the study area before, except for S. mansoni, which persists even 10 years after the initial report [24] and six rounds of mass deworming [88]. Only five of the 15 trematode species were identified to species level, highlighting the barcoding void. This barcoding void can only be resolved when adult trematode specimens are available, allowing distinct morphological features and life history traits to be linked to genetic information, as demonstrated by Schols et al. [28] and Laidemitt et al. [89]. Although we did not have access to such material, future studies can improve on this and already benefit from our newly established link between the trematode and intermediate host snail–information crucial for completing the parasite's life cycle.
While our trematode diversity aligns with local and regional studies, such as Schols et al. [30] reporting 19 species in three Zimbabwean districts and Outa et al. [90] documenting 17 species in Lake Victoria, Kenya, other regions exhibit even higher diversity. For instance, Lake Takvatn in the sub-Arctic region of Norway reported 24 trematode species [91], and a German nature reserve, Bienener Altrhein, documented 40 species [48]. However, sampling design and efforts differ between these studies, hindering meaningful comparisons.
Numerous factors contribute to trematode distribution, among which is the availability of suitable host snails. Our investigation uncovered notable parasite diversity variations between Wedza and Chiredzi, despite nearly identical snail host diversity as discussed above. Of the 15 trematode species, six were exclusive to Chiredzi, eight to Wedza, and only one species was shared between the two districts. The divergence in parasite transmission patterns in locations with comparable host species diversity is intriguing and highlights the complex factors contributing to geographical heterogeneity, including the widely known influence of aquatic environmental variables [92] and final vertebrate diversity [93]. Bulinus tropicus, responsible for the highest trematode diversity (five distinct trematode taxa), was exclusively found in Wedza. It is not uncommon for a single snail species to contribute significantly to trematode diversity. For example, the snail host Lymnaea stagnalis exhibited the highest prevalence and diversity, representing 47.6% of species richness across 21 freshwater lakes in Denmark [47]. Our findings, however, contrast with those of Babbitt et al. [94] who reported higher overall trematode diversity among B. globosus, B. ugandae, and B. forskalii (a minimum of five trematode taxa per species) compared to B. truncatus/tropicus specimens (two trematode taxa) in the Lake Victoria Basin of Kenya.
Conclusions
Our study reveals a high snail and trematode species diversity in Chiredzi and Wedza, but the identification of the trematode taxa to species level was seriously hampered by the lack of reference sequences in online databases. The distinct trematode communities encountered across geographic regions with similar freshwater snail hosts, highlights the necessity to explore the effects of other factors involved in disease transmission, including ambient environmental variables and the diversity of final hosts. Finally, the identification of important exotic invasive freshwater species, such as T. granifera (a new record in Zimbabwe), P. columella, and P. acuta, raises concerns about potential alterations in local trematode transmission dynamics, warranting further investigation.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and the supplementary materials. Data concerning DNA sequences used in this study are available on NCBI GenBank, with accession codes PP468524-PP468533, PP481879-PP481891, PP556550-PP556562, and PP564875-PP564882. Further inquiries or requests can be directed to the corresponding author.
References
Lai YS, Biedermann P, Ekpo UF, Garba A, Mathieu E, Midzi N, et al. Spatial distribution of schistosomiasis and treatment needs in sub-Saharan Africa: a systematic review and geostatistical analysis. Lancet Infect Dis. 2015;15:927–40.
WHO. Ending the neglect to attain the Sustainable Development Goals: a road map for neglected tropical diseases 2021–2030. Geneva, Switzerland: World Health Organization.; 2020. https://www.who.int/publications-detail-redirect/9789240010352. Accessed 15 Nov 2023.
Dey AR, Begum N, Anisuzzaman J, Islam MT, Alam MZ. A large-scale epidemiological investigation on trematode infections in small ruminants in Bangladesh. Vet Med Sci. 2022;8:1219–28.
Mehmood K, Zhang H, Sabir AJ, Abbas RZ, Ijaz M, Durrani AZ, et al. A review on epidemiology, global prevalence and economical losses of fasciolosis in ruminants. Microb Pathog. 2017;109:253–62.
Herrmann KK, Sorensen RE. Seasonal dynamics of two mortality-related trematodes using an introduced snail. J Parasitol. 2009;95:823–8.
Huffman JE, Roscoe DE. Experimental infections of waterfowl with Sphaeridiotrema globulus (Digenea). J Wildl Dis. 1989;25:143–6.
Mas-Coma MS, Esteban JG, Bargues MD. Epidemiology of human fascioliasis: a review and proposed new classification. Bull World Health Organ. 1999;77:340–6.
Parkinson M, O’Neill SM, Dalton JP. Endemic human fasciolosis in the Bolivian Altiplano. Epidemiol Infect. 2007;135:669–74.
Ashrafi K, Bargues MD, O’Neill S, Mas-Coma S. Fascioliasis: a worldwide parasitic disease of importance in travel medicine. Travel Med Infect Dis. 2014;12:636–49.
Le TH, De NV, Agatsuma T, Thi Nguyen TG, Nguyen QD, McManus DP, et al. Human fascioliasis and the presence of hybrid/introgressed forms of Fasciola hepatica and Fasciola gigantica in Vietnam. Int J Parasitol. 2008;38:725–30.
Nguyen TBN, Van De N, Nguyen TKL, Quang HH, Doan HTT, Agatsuma T, et al. Distribution status of hybrid types in large liver flukes, Fasciola species (Digenea: Fasciolidae), from ruminants and humans in Vietnam. Korean J Parasitol. 2018;56:453–61.
Huyse T, Webster BL, Geldof S, Stothard JR, Diaw OT, Polman K, et al. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLOS Pathog. 2009;5:e1000571.
Léger E, Borlase A, Fall CB, Diouf ND, Diop SD, Yasenev L, et al. Prevalence and distribution of schistosomiasis in human, livestock, and snail populations in northern Senegal: a one health epidemiological study of a multi-host system. Lancet Planet Health. 2020;4:e330-42.
Savassi BAES, Dobigny G, Etougbétché JR, Avocegan TT, Quinsou FT, Gauthier P, et al. Mastomys natalensis (Smith, 1834) as a natural host for Schistosoma haematobium (Bilharz, 1852) Weinland, 1858 x Schistosoma bovis Sonsino, 1876 introgressive hybrids. Parasitol Res. 2021;120:1755–70.
Díaz AV, Walker M, Webster JP. Reaching the World Health Organization elimination targets for schistosomiasis: the importance of a One Health perspective. Philos Trans R Soc B Biol Sci. 1887;2023:20220274.
Douchet P, Gourbal B, Loker ES, Rey O. Schistosoma transmission: scaling-up competence from hosts to ecosystems. Trends Parasitol. 2023;39:563–74.
Gower CM, Vince L, Webster JP. Should we be treating animal schistosomiasis in Africa? The need for a One Health economic evaluation of schistosomiasis control in people and their livestock. Trans R Soc Trop Med Hyg. 2017;111:244–7.
Chandiwana SK, Taylor P, Chimbari M, Ndhlovu P, Makura O, Bradley M, et al. Control of schistosomiasis transmission in newly established smallholder irrigation schemes. Trans R Soc Trop Med Hyg. 1988;82:874–80.
Mucheka VT, Lamb JM, Pfukenyi DM, Mukaratirwa S. DNA sequence analyses reveal co-occurrence of novel haplotypes of Fasciola gigantica with F. hepatica in South Africa and Zimbabwe. Vet Parasitol. 2015;214:144–51.
Pfukenyi DM, Mukaratirwa S, Willingham AL, Monrad J. Epidemiological studies of Fasciola gigantica infections in cattle in the highveld and lowveld communal grazing areas of Zimbabwe. Onderstepoort J Vet Res. 2006;73:37–51.
Pfukenyi DM, Mukaratirwa S. Amphistome infections in domestic and wild ruminants in East and Southern Africa: a review. Onderstepoort J Vet Res. 2018;85:e1-13.
Shiff CJ, Coutts WC, Yiannakis C, Holmes RW. Seasonal patterns in the transmission of Schistosoma haematobium in Rhodesia, and its control by winter application of molluscicide. Trans R Soc Trop Med Hyg. 1979;73:375–80.
Taylor P, Makura O. Prevalence and distribution of schistosomiasis in Zimbabwe. Ann Trop Med Parasitol. 1985;79:287–99.
Midzi N, Mduluza T, Chimbari MJ, Tshuma C, Charimari L, Mhlanga G, et al. Distribution of schistosomiasis and soil transmitted helminthiasis in Zimbabwe: towards a national plan of action for control and elimination. PLoS Negl Trop Dis. 2014;8:e3014.
Mutsaka-Makuvaza MJ, Matsena-Zingoni Z, Katsidzira A, Tshuma C, Chinombe N, Zhou XN, et al. Urogenital schistosomiasis and risk factors of infection in mothers and preschool children in an endemic district in Zimbabwe. Parasit Vectors. 2019;12:427.
Pantelouris EM. The common liver fluke, Fasciola hepatica L. New York: Pergamon Press; 1965.
Pfukenyi DM, Mukaratirwa S. A retrospective study of the prevalence and seasonal variation of Fasciola gigantica in cattle slaughtered in the major abattoirs of Zimbabwe between 1990 and 1999. Onderstepoort J Vet Res. 2004;71:181–7.
Schols R, Carolus H, Hammoud C, Muzarabani KC, Barson M, Huyse T. Invasive snails, parasite spillback, and potential parasite spillover drive parasitic diseases of Hippopotamus amphibius in artificial lakes of Zimbabwe. BMC Biol. 2021;19:160.
Carolus H, Muzarabani KC, Hammoud C, Schols R, Volckaert FAM, Barson M, et al. A cascade of biological invasions and parasite spillback in man-made Lake Kariba. Sci Total Environ. 2019;659:1283–92.
Schols R, Mudavanhu A, Carolus H, Hammoud C, Muzarabani KC, Barson M, et al. Exposing the barcoding void: an integrative approach to study snail-borne parasites in a one health context. Front Vet Sci. 2020;7:605280.
Muzarabani KC, Carolus H, Schols R, Hammoud C, Barson M, Huyse T. An update on snail and trematode communities in the Sanyati Basin of Lake Kariba: New snail and trematode species but no human schistosomes. Parasitol Int. 2024;99:102830.
Mugandani R, Wuta M, Makarau A, Chipindu B. Reclassification of agro-ecological regions of Zimbabwe in conformity with climate variability and change. Afr Crop Sci J. 2012;20:361–9.
Mandahl-Barth G. Key to the identification of east and central African freshwater snails of medical and veterinary importance. Bull World Health Organ. 1962;27:135–50.
Brown DS. Freshwater snails of Africa and their medical importance. 2nd ed. Vol. 75. UK: Taylor & Francis Ltd; 1994.
Appleton C, Miranda N. Two Asian freshwater snails newly introduced into South Africa and an analysis of alien species reported to date. Afr Invertebr. 2015;56:1–17.
Gumble A, Otori Y, Ritchie LS, Hunter GW. The effect of light, temperature and pH on the emergence of Schistosoma japonicum cercariae from Oncomelania nosophora. Trans Am Microsc Soc. 1957;76:87–92.
Frandsen F, Christensen NO. An introductory guide to the identification of cercariae from African freshwater snails with special reference to cercariae of trematode species of medical and veterinary importance. Acta Trop. 1984;41:181–202.
Ziętara MS, Arndt A, Geets A, Hellemans B, Filip A, Volckaert M. The nuclear rDNA region of Gyrodactylus arcuatus and G. branchicus (Monogenea: Gyrodactylidae). J Parasitol. 2000;86:1368–73.
Schols R, Carolus H, Hammoud C, Mulero S, Mudavanhu A, Huyse T. A rapid diagnostic multiplex PCR approach for xenomonitoring of human and animal schistosomiasis in a “One Health” context. Trans R Soc Trop Med Hyg. 2019;113:722–9.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6.
Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, et al. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47:W260-5.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 gateway computing environments workshop (GCE). 2010. p. 1–8.
Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformat. 2010;11:7.
Nwoko OE, Manyangadze T, Chimbari MJ. Spatial and seasonal distribution of human schistosomiasis intermediate host snails and their interactions with other freshwater snails in 7 districts of KwaZulu-Natal province, South Africa. Sci Rep. 2023;13:7845.
Mereta ST, Bedewi J, Yewhalaw D, Mandefro B, Abdie Y, Tegegne D, et al. Environmental determinants of distribution of freshwater snails and trematode infection in the Omo Gibe River Basin, southwest Ethiopia. Infect Dis Poverty. 2019;8:93.
Siama A, Eteme Enama S, Kalmobe J, Abah S, Foutchou A, Njan Nloga AM. Abundance, distribution, and diversity of freshwater snail and prevalences of their infection by cercaria of Fasciola gigantica and Schistosoma spp. at Mayo-Vreck River, Far North Region of Cameroon. J Trop Med. 2023;2032:e9527349.
Duan Y, Al-Jubury A, Kania PW, Buchmann K. Trematode diversity reflecting the community structure of Danish freshwater systems: molecular clues. Parasit Vectors. 2021;14:43.
Schwelm J, Selbach C, Kremers J, Sures B. Rare inventory of trematode diversity in a protected natural reserve. Sci Rep. 2021;11:22066.
Ouma JH, Waithaka FT. Bulinus tropicus (Krauss, 1848) from Kenya found naturally infected with Schistosoma bovis. Ann Trop Med Parasitol. 1984;78:341–2.
Grabner DS, Mohamed FAMM, Nachev M, Méabed EMH, Sabry AHA, Sures B. Invasion biology meets parasitology: a case study of parasite spill-back with Egyptian Fasciola gigantica in the invasive snail Pseudosuccinea columella. PLoS ONE. 2014;9:e88537.
Nhiwatiwa T, Dalu T, Brendonck L. Impact of irrigation based sugarcane cultivation on the Chiredzi and Runde Rivers quality, Zimbabwe. Sci Total Environ. 2017;587–588:316–25.
Aditya G, Raut SK. Predation potential of the water bugs Sphaerodema rusticum on the sewage snails Physa acuta. Mem Inst Oswaldo Cruz. 2002;97:531–4.
Rasifudi L, Addo-Bediako A, Bal K, Swemmer TM. Distribution of benthic macroinvertebrates in the Selati River of the Olifants River System. South Africa Afr Entomol. 2018;26:398–406.
Hoover CM, Rumschlag SL, Strgar L, Arakala A, Gambhir M, de Leo GA, et al. Effects of agrochemical pollution on schistosomiasis transmission: a systematic review and modelling analysis. Lancet Planet Health. 2020;4:e280-91.
de Kock KN, Wolmarans CT. Invasive alien freshwater snail species in the Kruger National Park. South Africa Koedoe. 2008;50:49–53.
Pointier JP. Invading freshwater gastropods: some conflicting aspects for public health. Malacologia. 1999;41:403–11.
Pointier JP, David P, Jarne P. Biological invasions: the case of planorbid snails. J Helminthol. 2005;79:249–56.
Appleton C. Alien and invasive fresh water Gastropoda in South Africa. Afr J Aquat Sci. 2003;28:69–81.
Appleton C, Forbes AT, Demetriades NT. The occurrence, bionomics and potential impacts of the invasive freshwater snail Tarebia granifera (Lamarck, 1822) (Gastropoda: Thiaridae) in South Africa. Zool Meded. 2009;83:525–36.
Appleton C, Nadasan DS. First report of Tarebia granifera (Lamarck, 1816) (Gastropoda: Thiaridae) from Africa. J Molluscan Stud. 2002;68:399–402.
Abbot RT. A study of an intermediate snail host (Thiara granifera) of the oriental lung fluke (Paragonimus). Proc U S Natl Mus. 1952;102:71–116.
de Necker L, van Rooyen D, Gerber R, Brendonck L, Wepener V, Smit NJ. Effects of river regulation on aquatic invertebrate community composition: a comparative analysis in two southern African rivers. Ecol Evol. 2024;14:e10963.
Giboda M, Malek EA, Correa R. Human schistosomiasis in Puerto Rico: reduced prevalence rate and absence of Biomphalaria glabrata. Am J Trop Med Hyg. 1997;57:564–8.
Pointier JP, Giboda M. The case for biological control of snail Intermediate hosts of Schistosoma mansoni. Parasitol Today. 1999;15:395–7.
Miranda N, Perissinotto R. Effects of an alien invasive gastropod on native benthic assemblages in coastal lakes of the iSimangaliso Wetland Park. South Africa Afr Invertebr. 2014;55:209–28.
Gomez Perez J, Vargas M, Malek EA. Displacement of Biomphalaria glabrata by Thiara granifera under natural conditions in the Dominican Republic. Mem Inst Oswaldo Cruz. 1991;86:341–7.
Pointier JP, Jourdane J. Biological control of the snail hosts of schistosomiasis in areas of low transmission: the example of the Caribbean area. Acta Trop. 2000;77:53–60.
de Kock KN, Joubert PH, Pretorius SH. Geographical distribution and habitat preferences of the invader freshwater snail species Lymnaea columella (Mollusca: Gastropoda) in South Africa. Onderstepoort J Vet Res. 1989;56:271–5.
Lounnas M, Correa AC, Vázquez AA, Dia A, Escobar JS, Nicot A, et al. Self-fertilization, long-distance flash invasion and biogeography shape the population structure of Pseudosuccinea columella at the worldwide scale. Mol Ecol. 2017;26:887–903.
Malatji MP, Mukaratirwa S. Molecular detection of natural infection of Lymnaea (Pseudosuccinea) columella (Gastropoda: Lymnaeidae) with Fasciola gigantica (Digenea: Fasciolidae) from two provinces of South Africa. J Helminthol. 2019;94:e38.
Lawton SP, Allan F, Hayes PM, Smit NJ. DNA barcoding of the medically important freshwater snail Physa acuta reveals multiple invasion events into Africa. Acta Trop. 2018;188:86–92.
Albrecht C, Kroll O, Moreno Terrazas E, Wilke T. Invasion of ancient Lake Titicaca by the globally invasive Physa acuta (Gastropoda: Pulmonata: Hygrophila). Biol Invasions. 2009;11:1821–6.
De Kock KN, Wolmarans CT. Distribution and habitats of the alien invader freshwater snail Physa acuta in South Africa. Water SA. 2007;33:717–22.
Madsen H, Frandsen F. The spread of freshwater snails including those of medical and veterinary importance. Acta Trop. 1989;46:139–46.
Dobson M. Replacement of native freshwater snails by the exotic Physa acuta (Gastropoda: Physidae) in southern Mozambique; a possible control mechanism for schistosomiasis. Ann Trop Med Parasitol. 2004;98:543–8.
Gashaw F, Erko B, Teklehaymanot T, Habtesellasie R. Assessment of the potential of competitor snails and African catfish (Clarias gariepinus) as biocontrol agents against snail hosts transmitting schistosomiasis. Trans R Soc Trop Med Hyg. 2008;102:774–9.
Habib MR, Lv S, Rollinson D, Zhou XN. Invasion and dispersal of Biomphalaria species: increased vigilance needed to prevent the introduction and spread of schistosomiasis. Front Med. 2021;8:614797.
Mutsaka-Makuvaza MJ, Zhou XN, Tshuma C, Abe E, Manasa J, Manyangadze T, et al. Molecular diversity of Bulinus species in Madziwa area, Shamva district in Zimbabwe: implications for urogenital schistosomiasis transmission. Parasit Vectors. 2020;13:14.
Manyangadze T, Chimbari MJ, Rubaba O, Soko W, Mukaratirwa S. Spatial and seasonal distribution of Bulinus globosus and Biomphalaria pfeifferi in Ingwavuma, uMkhanyakude district, KwaZulu-Natal, South Africa: implications for schistosomiasis transmission at micro-geographical scale. Parasit Vectors. 2021;14:222.
Leiper RT. The entozoa of the hippopotamus. In: Proceedings of the Zoological Society of London. Wiley Online Library; 1910. p. 233–52.
Schols R, Huyse T. Fasciola nyanzae. Trends Parasitol. 2024.
Catalano S, Sène M, Diouf ND, Fall CB, Borlase A, Léger E, et al. Rodents as natural hosts of zoonotic Schistosoma species and hybrids: an epidemiological and evolutionary perspective from West Africa. J Infect Dis. 2018;218:429–33.
Miranda GS, Rodrigues JG, de Oliveira Silva JK, Camelo GM, Silva-Souza N, Neves RH, et al. New challenges for the control of human schistosomiasis: the possible impact of wild rodents in Schistosoma mansoni transmission. Acta Trop. 2022;236:106677.
Weyher AH, Phillips-Conroy JE, Fischer K, Weil GJ, Chansa W, Fischer PU. Molecular identification of Schistosoma mattheei from feces of Kinda (Papio cynocephalus kindae) and grayfoot baboons (Papio ursinus griseipes) in Zambia. J Parasitol. 2010;96:184–90.
Laidemitt MR, Anderson LC, Wearing HJ, Mutuku MW, Mkoji GM, Loker ES. Antagonism between parasites within snail hosts impacts the transmission of human schistosomiasis. Elife. 2019;8:e50095.
Beverly-Burton M. A new strigeid Diplostomum (Tylodelphys) mashonense n. sp. (Trematoda: Diplostomatidae) from the grey heron, Ardea cinerea L. in Southern Rhodesia, with an experimental demonstration of part of the life cycle. Rev Zool Bot Afr. 1963;68:291–308.
Barson M, Bray R, Ollevier F, Huyse T. Taxonomy and faunistics of the helminth parasites of Clarias gariepinus (Burchell, 1822), and Oreochromis mossambicus (Peters, 1852) from temporary pans and pools in the Save-Runde River floodplain, Zimbabwe. Comp Parasitol. 2008;75:228–40.
Mduluza T, Jones C, Osakunor DNM, Lim R, Kuebel JK, Phiri I, et al. Six rounds of annual praziquantel treatment during a national helminth control program significantly reduced schistosome infection and morbidity levels in a cohort of schoolchildren in Zimbabwe. PLoS Negl Trop Dis. 2020;14:e0008388.
Laidemitt MR, Zawadzki ET, Brant SV, Mutuku MW, Mkoji GM, Loker ES. Loads of trematodes: discovering hidden diversity of paramphistomoids in Kenyan ruminants. Parasitology. 2017;144:131–47.
Outa JO, Sattmann H, Köhsler M, Walochnik J, Jirsa F. Diversity of digenean trematode larvae in snails from Lake Victoria, Kenya: first reports and bioindicative aspects. Acta Trop. 2020;206:105437.
Soldánová M, Georgieva S, Roháčová J, Knudsen R, Kuhn JA, Henriksen EH, et al. Molecular analyses reveal high species diversity of trematodes in a sub-Arctic lake. Int J Parasitol. 2017;47:327–45.
Maes T, Hammoud C, Volckaert FAM, Huyse T. A call for standardised snail ecological studies to support schistosomiasis risk assessment and snail control efforts. Hydrobiologia. 2021;848:1773–93.
Hechinger RF, Lafferty KD. Host diversity begets parasite diversity: bird final hosts and trematodes in snail intermediate hosts. Proc R Soc B Biol Sci. 2005;272:1059–66.
Babbitt CR, Laidemitt MR, Mutuku MW, Oraro PO, Brant SV, Mkoji GM, et al. Bulinus snails in the Lake Victoria Basin in Kenya: Systematics and their role as hosts for schistosomes. PLoS Negl Trop Dis. 2023;17:e0010752.
Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford, New York: Oxford University Press; 2000. p. 348.
Hammoud C, Mulero S, Van Bocxlaer B, Boissier J, Verschuren D, Albrecht C, et al. Simultaneous genotyping of snails and infecting trematode parasites using high-throughput amplicon sequencing. Mol Ecol Resour. 2021;22:567–86.
Waeschenbach A, Webster BL, Littlewood DTJ. Adding resolution to ordinal level relationships of tapeworms (Platyhelminthes: Cestoda) with large fragments of mtDNA. Mol Phylogenet Evol. 2012;63:834–47.
Littlewood DTJ, Olson PD. Small subunit rDNA and the Platyhelminthes: Signal, noise, conflict and compromise. In: Interrelationships of the Platyhelminthes. CRC Press; 2001.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols. Elsevier; 1990. p. 315–22.
Acknowledgements
This study was made possible with logistical support received from the Malilangwe Trust in Chiredzi, especially through its director Mark Saunders, scientists Dr. Bruce Clegg (PhD), Sarah Clegg, and Allan Tarugara, and research technician Philemon Chivambu. Our thanks also go out to Reilly Travers of Imire Rhino and Wildlife Conservation for his warm welcome and assistance. A great deal of scientific help was provided by Bart Hellemans at the KU Leuven, Laboratory of Biodiversity and Evolutionary Genomics. We were granted access to study Hippo Valley Estates by Bekithemba Ndlovu, Faustina Zvenyika, and Dr. Tongai Mukwewa (MD), whose insights into local disease history were vital to our research. We thank Ruben Put for his hand in the field. The graphical abstract was created with BioRender.com.
Funding
AM was financially supported by the Vlaamse Interuniversitaire Raad (Flemish Interuniversity Council)–Universitaire Ontwikkelingssamenwerking (VLIR-UOS) through a Global Minds PhD Fellowship (Project no. 3E210232) and logistically supported by the Malilangwe Trust. RS was supported by BRAIN-be 2.0 under the MicroResist project (B2/191/P1/MicroResist). We acknowledge the travel grant from Stichting ter Bevordering van het Biodiversiteitsonderzoek in Afrika/Fondation pour Favoriser la Recherche sur la Biodiversité en Afrique (SBBOA/FFRBA) awarded to RS and AM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: AM, RS, LB, and TH; formal analysis: AM; investigation: AM, and RS; resources: LB and TH.; data curation: AM, and RS; writing–original draft: AM; writing–review and editing: AM, RS, EG, BC, TN, TM, LB, and TH; supervision: TN, TM, LB, and TH. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate.
The research permit for collecting freshwater snails and monitoring trematodes was granted by the National Animal Research Ethics Council of Zimbabwe (Contract No: 006/2020). There was a need to transfer specimens to Belgium for DNA analysis. As such, a memorandum of transfer was signed between Bindura University of Science Education, the University of Leuven, and the Royal Museum of Central Africa (RMCA) outlining proper use for research purposes and curation. To export the samples, we sought the permission of the National Biotechnology Authority of Zimbabwe (Permit No: BEP0009211, Reference No: NBA/RA/10/01–0921) in compliance with the Nagoya Protocol on Access and Benefit-sharing (https://www.cbd.int/abs/). Further, permission to import the material into Belgium was granted to RMCA by the Federal Agency for the Safety of the Food Chain (Reference No: 2021DBP083).
Consent for publication
Not applicable.
Competing interests
The authors affirm that the research was conducted in the absence of any commercial, financial and/or personal relationships that could be interpreted as a potential competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mudavanhu, A., Schols, R., Goossens, E. et al. One Health monitoring reveals invasive freshwater snail species, new records, and undescribed parasite diversity in Zimbabwe. Parasites Vectors 17, 234 (2024). https://doi.org/10.1186/s13071-024-06307-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-024-06307-4