Abstract
Background
Lactiplantibacillus plantarum HEAL9 and Lacticaseibacillus paracasei 8700:2 positively affect the fecal bacteriome in children with celiac disease autoimmunity after 6 months of supplementation. The aim of the present investigation was to study the effects of Lactiplantibacillus plantarum HEAL9 and Lacticaseibacillus paracasei 8700:2 on the single-cell parasitome, with a primary focus on Blastocystis.
Methods
Stool samples were collected from 78 Swedish children with celiac disease autoimmunity participating in a randomized, double-blind, placebo-controlled clinical trial to either receive a mixture of supplementation with L. plantarum HEAL9 and L. paracasei 8700:2 (n = 38) or placebo (n = 40). A total of 227 stool samples collected at baseline and after 3 and 6 months of intervention, respectively, were retrospectively analyzed for Blastocystis by quantitative real-time PCR and subtyped by massively parallel amplicon sequencing. Other single-cell parasites were detected by untargeted 18S rDNA amplicon sequencing and verified by real-time PCR. The relation between the parasites and the bacteriome community was characterized by using 16S rDNA profiling of the V3-V4 region.
Results
Three different single-cell protists were identified, of which the highest prevalence was found for Dientamoeba fragilis (23.1%, 18/78 children), followed by Blastocystis (15.4%, 12/78) and Entamoeba spp. (2.6%, 2/78). The quantity of the protists was stable over time and not affected by probiotic intervention (P = 0.14 for Blastocystis, P = 0.10 for D. fragilis). The positivity of the protists was associated with increased bacteriome diversity (measured by multiple indices, P < 0.03). Bacterial composition was influenced by the presence of the protists: positivity of Blastocystis was inversely associated with Akkermansia (at the levels of the genus as well as its family, order, class and phylum); P < 0.002), Faecalibacterium (P = 0.003) and Romboutsia (P = 0.029); positivity of D. fragilis was inversely associated with families Enterobacteriaceae (P = 0.016) and Coriobacteriaceae (P = 0.022) and genera Flavonifractor (P < 0.001), Faecalibacterium (P = 0.009), Lachnoclostridium (P = 0.029), Ruminococcus (P < 0.001) and Granulicatella (P = 0.018).
Conclusions
The prevalence of single-cell protists is low in children with celiac disease autoimmunity. The colonization was stable regardless of the probiotic intervention and associated with increased diversity of the fecal bacteriome but inversely associated with some beneficial bacteria.
Graphical Abstract

Similar content being viewed by others
Background
The main protist of the human gut is Blastocystis sp. (hereafter referred to by its genus name Blastocystis). It is an anaerobic, non-flagellated single-cell and highly polymorphic organism with multiple forms. Blastocystis is proposed to be a marker of a balanced microbiome [1,2,3] and is believed to be the most common single-cell eukaryote in the gut of both adults [4] and children [5]. Being mainly transferred by the fecal-oral route, Blastocystis has a higher prevalence in developing countries with lower hygiene standards, usually between 40 and 84% [6,7,8], compared with high-income countries in Europe and the USA, where it commonly ranges between 7 and 56% [1, 9,10,11,12,13,14,15]. Among the genus Blastocystis, 26 genetic subtypes (ST) were identified [16] and proposed to be differing in biological properties. Of these subtypes, STs 1–17 are widely recognized as valid, whereas others may include molecular detection artifacts [17]. Of the ten subtypes in humans, ST1 to ST4 are the most prevalent [5, 18], from which ST4 is unique for Europe and ST3 is the most prevalent worldwide [10]. Another frequent protist is Dientamoeba fragilis, a flagellated trichomonad, surrounded by controversy about whether it is more a pathogen or a benign gut inhabitant [19,20,21]. Its prevalence varies between 0.4 and 71%, depending on the studied cohort and methods used [21,22,23,24]. Interestingly, unlike Blastocystis, its prevalence is lower in low-income countries, e.g. [25, 26], and higher in high-income countries, e.g. [22, 27].
Celiac disease is a chronic enteropathy due to an immune-mediated response to dietary gluten from wheat, rye and barley, arising in a small proportion of individuals who are genetically susceptible because carrying human leukocyte antigens (HLA) haplotypes DQ2 and/or DQ8 [28]. Although genetics and gluten are necessary for celiac disease development, it is most likely triggered by environmental factors such as gastrointestinal infectious episodes triggered by different microorganisms [29], 29. On the other hand, the prevalence of celiac disease has been reported to be significantly lower in populations of Russian Karelia with lower hygienic and socioeconomic standards with high exposures to microbes compared to populations living in Finland, despite the populations from the two geographically neighboring regions sharing the same genetic risk [31].
In contrast to the above study, recent prospective birth cohort studies on gastrointestinal infections have shown that patients who develop celiac disease have more frequent enterovirus [32] and parechovirus [33] infections prior to seroconversion of tissue transglutaminase autoantibodies (tTGA), a marker of celiac disease autoimmunity (CDA) [32].
The present study builds on the probiotic intervention trial in children with CDA [34], showing a moderate effect of probiotics on the gut bacteriome [35]. This study focuses on the fecal parasitome. The aim was to assess the effect of the intervention with two lactobacilli, probiotic strains Lactiplantibacillus plantarum HEAL9 and Lacticaseibacillus paracasei 8700:2, on the single-cell parasitome with specific emphasis on Blastocystis in a stool sample set of Swedish children with CDA. Our hypothesis was that the presumably beneficial probiotic bacteria might change the bacteriome composition, leading to an increased prevalence of Blastocystis.
Materials and methods
Study population
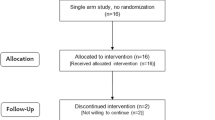
The Celiac Disease Prevention with Probiotics (CiPP) study, a double-blind placebo-controlled randomized clinical trial, was performed at the Department of Clinical Sciences, Unit of Celiac Disease and Diabetes, Lund University, Malmö, Sweden [34]. The trial was carried out between March 12, 2012, and August 25, 2015. A total of 118 children with CDA identified by screening were invited to participate in the CiPP study, of whom 89 accepted participation and 78 completed a 6-month follow-up of daily receiving either a mixture of L. plantarum HEAL9 and L. paracasei 8700:2 (probiotic group, n = 40) or 1 g maltodextrin and yeast peptone (placebo group, n = 38) (the probiotic mixture is described in more detail in [34, 35]). Study participants were instructed to continue a gluten-containing diet and exclude other food products containing probiotics. During the intervention, six children in the probiotic group and four in the placebo group reported taking antibiotics (P = 0.561). Stool samples were collected at baseline and then 3 and 6 months after the intervention. Processing of the stool samples for DNA extraction was described earlier [35]. Briefly, 50 mg of thawed stool sample was used for the DNA extraction using the DNA tissue kit (Qiagen, Germany) and run on the EZ1 DNA extraction robot (Qiagen, Germany). The study was approved by the Ethics Committee of the Medical Faculty, Lund University (Dnr 2011/335; Dnr 2021–04470) and registered at ClinicalTrials.gov (NCT03176095); 227 samples collected from the 78 children with CDA participating in the (CiPP) study were available for this retrospective analysis (Fig. 1).
Molecular methods
Blastocystis was tested and quantified by specific real-time PCR as the most common gut protist [18]. All positive samples for Blastocystis were then classified into genetic subtypes using massively parallel amplicon sequencing of a unique region of the 18S rDNA gene. The total parasitome was profiled by multiplex massively parallel amplicon sequencing of several amplicons of the 18S rDNA gene. Positive parasites from the 18S rDNA profiling (other than Blastocystis) were then confirmed and quantified by parasite-specific real-time PCR. After determining the single-cell parasitome content, the parasites were related to the intervention with studied probiotics and analyzed in the context of previously obtained bacteriome profiles [35].
Detection and quantification of blastocystis by specific real-time quantitative PCR
A real-time quantitative PCR assay with a specific probe for Blastocystis designed by Stensvold et al. [36] was used to test and quantify Blastocystis. For the calibration curve, DNA from a microscopically quantified xenic culture was used [37]. Platinum Taq Polymerase (Invitrogen, USA) from the Stensvold’s original protocol was substituted by the HotStar Taq polymerase chemistry (Qiagen, Germany), and the PCR program was corrected accordingly with 15 min of initial denaturation at 95 °C followed by 45 cycles of 15 s denaturation at 94 °C and 60 s of combined annealing and synthesis at 60 °C [5]. Reactions were performed in duplicates throughout the study, and negative controls were included in the extraction as well as in the detection reactions. PCR tubes containing real-time PCR products were always discarded unopened. The detection PCR fragments do not overlap with those used for Blastocystis subtyping.
Subtyping of blastocystis by massively parallel amplicon sequencing
Samples positive for Blastocystis in the above-mentioned real-time PCR assay were thereafter subtyped by massively parallel amplicon sequencing using the protocol by Maloney et al. [38] with primers developed by Santin et al. [39] extended by Nextera tails. The amplicons were then purified and indexed by Nextera XT Index Kit v2 Set A and D (Illumina, USA) by a short eight-cycle PCR, enabling multiplex sequencing. Indexed products were purified and equalized using a SequalPrep Normalization plate (Invitrogen, USA) and pooled. The pool of libraries was sequenced on a MiSeq instrument using Reagent Kit v2, 2 × 250 bp (Illumina, USA), with the addition of 20% PhiX control to balance the amplicon signal.
Parasitome survey using massively parallel amplicon profiling of the 18S rDNA gene
The parasitome survey was performed using massively parallel sequencing with five different PCR amplicons of the 18S rDNA gene, as reported in detail previously [5]. The procedure identified 3546 unique sequence variants and zero-radius operational taxonomic units (ZOTUs); those with a relative frequency > 0.1% (n = 103) were manually classified by BLAST search to GenBank. Signals of the single-cell protists were expressed as the number of reads aligned to a particular organism divided by the count of rarefied reads per reaction.
Quantification of D. fragilis and Entamoeba spp. by quantitative real-time PCR
The single-cell protists with a signal in the parasitome survey (D. fragilis and Entamoeba spp. apart from already quantified Blastocystis) were then quantified by a specific probe-based real-time quantitative PCR assay. For D. fragilis, the primer and probe sequences came from the work of Verweij et al. [40]. A dilution series of DNA from a xenic culture of D. fragilis was used as a control in qPCR reactions, but its exact protist genomic equivalent content could not be established because of problems with the microscopic quantification of the culture. The PCR conditions for D. fragilis detection were published in [41]. As to Entamoeba spp. (E. histolytica, dispar, moshkovskii, hartmanni, coli), the primer and probe sequences were designed in the Geneious Prime software (version 2020) based on the 18S SSU rRNA gene sequence references (X64142, AB197936, Z49256, AF149906, AF149907, AF149915) retrieved from the GenBank database (see Additional file 2: Table S1 and Figure S1). The PCR program for Entamoeba spp. detection consisted of 15 min of initial denaturation at 95 °C followed by 35 cycles of 15 s denaturation at 94 °C and 60 s of combined annealing and synthesis at 60 °C. For a calibration curve, DNA from an axenic Entamoeba culture was used, and cell counts from the culture were calculated using a Bürker chamber and then serially diluted to obtain aliquots containing 1, 10, 102, 103, 104 and 105 cells per microliter, which were subsequently subjected to DNA extraction according to Lhotská et al. [15]. Negative controls were included in every PCR run, and the tubes with PCR products were discarded unopened.
Bacteriome analysis
The processing, DNA library preparation and sequencing had been done previously by massively parallel amplicon sequencing of the V3-V4 region of the bacterial 16S rRNA gene [35]. For this study, the previously generated fastq files with sequencing reads were downloaded and reprocessed using the DADA2 pipeline (version 1.22) [42], with taxonomic classification using the SILVA database (version 138) [43] instead of the now slightly outdated Greengenes database version 13.5 and the Qiime2 suite. Amplicon sequence variants (ASVs, analogous to operational taxonomic units, OTUs) were then further analyzed using vegan [44] and phyloseq [45] in the R programming language [46].
Statistical analysis
The prevalence of single-cell protists was determined by counting any PCR positivity at any level. Their presence and quantity were then modeled using generalized estimating equations (GEE) with the subject as the grouping variable, time point (factor with three levels—0, 3 and 6 months) and a first-order autoregressive covariance structure; the predictors were the study allocation (placebo or probiotics) and whether the sample was taken while on intervention with the live mixture.
Bacteriome alpha (within-sample) diversity was assessed from the unfiltered rarefied dataset, agglomerated at the genus level, using the observed counts, Chao1, ACE, Shannon, Simpson, inverse Simpson and Fisher indices. The association of alpha diversity indices with the presence or quantity of Blastocystis or D. fragilis was assessed in GEE models with the index as the dependent variable where predictors were the positivity or quantity of the parasites, and the study allocation (placebo or probiotics).
For the beta diversity (between samples) analysis, Bray-Curtis distance was calculated on Hellinger-transformed abundance data agglomerated at the taxonomic level of the genus, and ordination was performed using metric multi-dimensional scaling (MDS) and visually inspected. Associations of Blastocystis and/or D. fragilis positivity with fecal bacteriome composition were tested using constrained ordination, the redundancy analysis (RDA) on the Hellinger-transformed abundance data.
Using GEE, individual bacterial taxa were tested for associations with the positivity of the two abundant protists (Blastocystis or D. fragilis). Taxa having more than 20/15000 reads in at least two samples were considered. In this model, the outcome was the relative abundance of a bacterium, and the predictors were positivity for Blastocystis and/or Dientamoeba, time point (factor with three levels—0, 3 and 6 months) and intervention (on probiotics vs. on placebo). An autoregressive correlation matrix was used. The models were built for every taxonomic unit at the genus, family, order, class and phylum levels. Ensuing nominal P values were adjusted using the Bonferroni method for the number of taxonomic entities tested at the given level. In an additional model, the dependent variable was the fold difference of bacterium quantity from the ASV table after the centered log-ratio transformation (CLR) with a small pseudo-count replacing zero values.
The commented R Markdown code and output of the statistical analysis are shown, along with the session information, as Additional file 1: Statistical analysis—R Markdown.
Results
Fecal parasitome analysis
The molecular survey using 18S rDNA parasitome profiling and/or specific quantitative PCR for the single-cell protists revealed positivity in widely varying quantities: in Blastocystis the threshold cycles (Ct) ranged from 17.9 to 41.2, corresponding to quantities of < 1/100 to > 10,000,000 genomic equivalents (g.e.) per µl DNA. In D. fragilis, the threshold cycles ranged from 19.6 to 42.6; the abundance in genomic equivalents is not known since the standard culture could not be microscopically quantified. In Entamoeba, the threshold cycles ranged from 23.2 to 30.3, corresponding to quantities of 1 to > 100 genomic equivalents per µl DNA.
The subject-wise prevalence of these three protists, calculated as at least once positive among the three time points, was highest in D. fragilis (18/78, 23.1%), slightly lower in Blastocystis (12/78, 15.4%) and the lowest in Entamoeba spp. (2/78, 2.6%) (Table 1 and Additional file 2: Fig. S2). Dientamoeba fragilis was more frequent in Blastocystis-positive samples (P = 0.015) but not the other way around (P = 0.057, both from GEE models). Entamoeba was only present in stable loads in one subject in each intervention group.
The protist frequency did not differ between the treatment groups (intervention vs. placebo) at baseline, and no significant effect on protist frequency was noted upon receiving the probiotic intervention (GEE with terms for the allocated treatment group, studied predictor being the ongoing intervention with lactobacilli: P = 0.34 for positivity of Blastocystis; P = 0.14 for D. fragilis; P = 0.31 for any of the three protists. Similarly, no influence of the intervention was noted on protist quantity—this was assessed in GEE models with the logarithm of quantity as the outcome (Fig. 2).
Blastocystis subtypes
Of 26 Blastocystis-positive samples, 25 had their subtype identified, while one failed in the sequencing (likely because of very low quantity, Ct 41.2). Four Blastocystis subtypes were identified: the most prevalent one was ST2 (10/25 identified, 40%), followed by ST4 (7/25, 28%), ST1 (5/25, 20%) and ST3 (3/25, 12%). None of the samples was positive for more than one subtype (Fig. 3A and B). Quantity did not differ by Blastocystis sequence type (P = 0.46), and there was no difference in the subject-wise prevalence of the four observed subtypes. The overview of each sample’s results (qPCR, subtyping and parasitome survey) is summarized in the Additional file 3: Table—Parasites detection.
Protists and the fecal bacteriome
The presence of Blastocystis and/or D. fragilis was associated with a higher alpha diversity of the fecal bacteriome. This association was significant in a GEE model for the count of observed genera (P = 0.018), the Chao1 (P = 0.013), ACE (P = 0.014), Shannon (P = 0.023) and Fischer (P = 0.017) indices as well as for the Simpson (P = 0.0057) and inversed Simpson (P = 0.026) indices (Fig. 4). The difference in alpha diversity between protist-positive and -negative stools was most prominent in those collected at the second time point. Alpha diversity neither changed over time nor differed between treatment groups at baseline and was not associated with the probiotic intervention.
Redundancy analysis performed on Hellinger-transformed bacterial abundance data indicated no effect of the intervention (P = 0.591), but the bacteriome community composition was associated with the presence of Blastocystis (P = 0.006) and D. fragilis (P = 0.001) (Additional file 2: Figure S3). The proportion of overall community variance explained by the two protists was low (1.0% for Blastocystis and 1.95% for D. fragilis). The effects of the two protozoa were nearly orthogonal, i.e. the associations with individual microbes of the bacteriome differed. Also, upon an inspection of ordination plots of Bray–Curtis distance at the genus level, samples positive for either Blastocystis and/or D. fragilis showed a tendency towards moderate separation from those being negative for the parasites (Additional file 2: Figure S4). As the spread of the two categories significantly differed (function vegan::betadisper, P < 0.001), testing by permutational analysis of variance (PERMANOVA) was not meaningful—its significance (P < 0.001) reflected either the difference in centroid position or the above-demonstrated difference in the spread, or both.
When studying the bacterial taxa associated with positivity for parasites using adjusted GEE (Table 2), Blastocystis was inversely associated with the genera Akkermansia (and its taxonomic categories up to the level of phylum Verrucomicrobia), Faecalibacterium and Romboutsia, both from class Clostridia. The presence of D. fragilis was inversely associated with the genera Flavonifractor, Faecalibacterium, Lachnoclostridium, Ruminococcus and Granulicatella. Most of the above associations were apparent in both the linear and fold-difference models, except for Akkermansia, whose association was detectable only when linear quantity was used as the outcome.
Discussion
This study analyzed the single-cell parasitome in a randomized clinical trial of probiotic strain intervention in CDA and found a relatively stable protist colonization (Blastocystis and/or D. fragilis) regardless of the probiotic intervention. The intervention with the two lactobacilli caused moderate changes in the bacteriome [35]; however, we observed no effects on the parasitome composition. Rather, the opposite occurred: positivity for a parasite was stable over the observation period and associated with an increased richness of the bacteriome but inversely associated with some presumably beneficial bacteria.
By targeted molecular testing using quantitative real-time PCR, we first looked at the prevalence of Blastocystis as the prime representative of the fecal parasitome [4, 5]. Then, we explored the whole parasitome by 18S rDNA profiling and assessed the quantities of the positive ones by specific real-time PCR assays for each of the protists. The subject-wise prevalence of the single-cell protists among children with CDA was low (23.1% for D. fragilis, 15.4% for Blastocystis and only 2.6% for Entamoeba spp.), in accordance with the hygiene hypothesis [47] and also to a recent report in celiac disease patients (regarding Blastocystis) [48].
The prevalence of Blastocystis is reported to be higher in lower-income countries and vice versa. Among the high-income countries, the lowest prevalence in a healthy population of only 7% was reported in Colorado, USA [13]. In continental Europe, it ranges around 20–30% (France 18%, The Netherlands 24%, Czechia 24%, Belgium 30%) [1, 11, 12, 15] and surprisingly higher prevalence of 56% was reported in Ireland [14]. On the other hand, two studies from rural populations of Nigeria [6] and Senegal [49] report a very high prevalence of 84% or 100%, respectively. Similarly, in underprivileged areas of Malaysia and Brazil, a higher prevalence of 41% and 47%, respectively, was reported [7, 8]. Thus, our results are in accord with the Blastocystis geographical gradient in healthy adults and children.
Although Blastocystis is often reported as the most common gut eukaryote [4], D. fragilis was more prevalent in the present dataset. This might be due to its higher prevalence in developed, urbanized countries [22, 27], but perhaps the location in the south of Sweden played a more prominent role, as supported by Jokelainen, who reported high prevalence in asymptomatic children under the age of 6 in day-care centeres in Copenhagen [20]. Additionally, a high prevalence of D. fragilis (compared to other intestinal parasites) was reported in urban areas of Copenhagen, Denmark [22, 50] (in adults) and Jönköping, Sweden [51] (in children) among patients with gastrointestinal issues suspected of parasitosis. Thus, the higher rates of D. fragilis than Blastocystis in our study seem meaningful regarding the geographic proximity to Copenhagen. Still, the high prevalence of D. fragilis in the region raises the question of its origin. It might be an endemic issue, but it might also be a simple reporting bias caused by the existence of several well-established parasite research groups in Denmark and Sweden. Similar isolated cases of a surprisingly high prevalence of Dientamoeba have been recently reported, e.g. in The Netherlands [52, 53] and Czechia [21], where groups studying single-cell protists also utilized molecular detection.
The colonization by these protists was stable over time and not influenced by the intervention of lactobacilli strains. The temporal stability in children was well described for D. fragilis [20]; however, for Blastocystis there is only one study in adults with a small number of subjects (n = 10) [14], making our study the first to report such stability of the parasitome among children.
The mixture of two lactobacilli strains was found to modulate the immune response [34] and change the fecal bacteriome towards a healthier composition [35]. However, it showed no detectable effect in modulating the fecal parasitome in presumably asymptomatic CDA patients. In contrast, probiotics may modulate parasite positivity in symptomatic patients: two studies investigated the effects of probiotics in treating symptomatic Blastocystis hominis infection. Dinleyici et al. showed a significant effect of Saccharomyces boulardii probiotics on Blastocystis eradication similar to metronidazole in vivo [54]; Lepczynska et al. reported an effect of a mixture of probiotics (Lactobacillus rhamnosus, Lactococcus lactis and Enterococcus faecium) on Blastocystis ST3 eradication in vitro [55].
In our study, ST2 was the most often identified subtype among study samples, albeit ST3 is reported as the most frequent worldwide [18]. ST3 was, on the contrary, found to be the least frequent. The occurrence of ST4, even as the second most common, is consistent with its strict European predilection [4]. Otherwise, the subtype distribution showed no association with the intervention.
The fecal bacteriome reaction to the mixture of L. plantarum HEAL9 and L. paracasei 8700:2 in our RCT was described earlier[35]. Briefly, the 6-month intervention with probiotics led to a shift in bacteriome composition in a healthier direction, most notably to an increase in Akkermansia, however, without any changes in alpha diversity indices. Celiac disease itself has been associated with changes in the gut bacteriome [56,57,58,59,60]. More specifically, it was associated with an increase in pro-inflammatory bacteria Enterobacteriaceae[56], Bacteroides [59] and Fusobacterium [56] and a decrease in Akkermansia [56], a beneficial bacterium for the gut epithelium [61]. Moreover, potential predictors of celiac disease were suggested, including an increase in Porhyromonas, Dialister and Parabacteroides and a decreased abundance of anti-inflammatory species [60] or decreased capacity to degrade gluten [57]. Importantly, 3-month probiotic supplementation shifted the bacteriome towards a healthier composition [58]. In contrast, recent work, with low patient count but innovative design, provided evidence that celiac disease might not be consistently associated with dysbiotic microbiome[62].
The probiotics did not influence the bacteriome’s alpha diversity; however, we found evidence of protist positivity increasing the alpha diversity similar to what has previously been described in adults, e.g. [2], and children [5]. As speculated in our previous work, the relation between protist positivity and rich bacteriome ecosystem might be in the thriving of Blastocystis in distinct communities [5] or the protist itself modifying the ecosystem [63]. Moreover, based on our current findings, D. fragilis, not just Blastocystis, is associated with fecal bacteriome diversity.
The positivity of Blastocystis and/or D. fragilis was associated with a difference in bacterial community composition. Even though the protist occurrence explained only a small part of the bacteriome beta diversity, the association was highly significant. Of particular taxa associated with Blastocystis, a negative association with Akkermansia and its upward taxonomic categories stands out. Akkermansia is a known mucin degrader and producer of short-chain fatty acids (SCFA) [64] and is thus generally considered beneficial to the human gut ecosystem [61]. It was found to be depleted in celiac disease [56] and, on the contrary, enriched after receiving probiotics in the CiPP study [35]. Of note, here, the Akkermansia association was only significant in the linear model, and the absolute difference is not high. We speculate that the bacterium does not thrive in an ecosystem engineered by Blastocystis. Another beneficial bacterium, Faecalibacterium, a known producer of SCFA [65] having anti-inflammatory properties [66], was also inversely associated with Blastocystis positivity. Romboutsia has been previously associated with an increased risk of nonalcoholic fatty liver disease [67] and neurodevelopmental disorders in children [68]; however, recent rigorous work found its beneficial role in cardiometabolic health [69]. All this combined makes its weak inverse association with Blastocystis ambiguous to interpret.
The presence of D. fragilis was, among others, inversely associated with Enterobacteriaceae, an inflammation-promoting bacteria known to be enriched in the bacteriomes of celiac disease patients [56]. On the other hand, its presence was inversely associated with many beneficial bacteria producing beneficial SCFA (including Flavonifractor, Faecalibacterium or Lachnospiraceae family), leaving the interpretation of these associations also complicated.
Strengths and limitations
Our study provides a unique insight into a novel topic in celiac disease research as no study has yet investigated the gut parasitome by molecular techniques. The samples were taken longitudinally, which helps decrease the effects of short-term fluctuations in the abundance of individual microbial taxa [70]. Analysis was performed by several complementary methods: Blastocystis detection by real-time PCR assay was followed by its subtyping using amplicon sequencing of another rDNA region; in remaining parasites the positivity in the parasitome survey by massive parallel sequencing was then confirmed using real-time PCR. This, along with the inclusion of multiple negative controls, safeguarded against false positivity. Furthermore, the present study extended previous investigations to explore the whole parasitome by using multiple primer pairs [5]. Moreover, we reported the quantity of the protists, not only dichotomous positivity in analyses of subtypes but also in association with the intervention. Although absolute quantification from stool is cumbersome (as there is no internal standard for reference), we still believe that this quantitative aspect adds confidence to our findings, even though we did not find any association with the protist quantity.
The main limitation is the unavailability of a fresh fecal sample needed for direct morphological assessment by microscopy as only frozen samples were used for DNA extraction. Thus, we do not know what stages of the protist were present and whether morphology relates to the quantity of Blastocystis subtypes. However, such an interpretation of the microscopy result might still be difficult, given that Blastocystis possesses one or two nuclei [71]. Another limitation is that no background population without CDA was investigated for protist prevalence comparison as the trial was designed far earlier than this parasitome study. Thus, no control group of healthy children without CDA was used for establishing protist prevalence in the background population.
Conclusions
The prevalence of Blastocystis and D. fragilis in children with CDA is rather low, with D. fragilis being the more prevalent of the two protists. Their positivity or quantity did not appreciably change upon the probiotic intervention. The presence of Blastocystis and D. fragilis was linked with an increased bacteriome diversity, although inversely associated with the abundance of some beneficial bacteria, like e.g. Akkermansia muciniphila. Even though the probiotics may help children with CDA to modulate the immune response and positively affect the fecal bacteriome, the single-cell parasitome remains unaffected.
Availability of data and materials
Data are available upon reasonable request.
Abbreviations
- CDA:
-
Celiac disease autoimmunity
- CiPP:
-
Celiac disease Prevention with Probiotics
- qPCR:
-
quantitative real-time PCR
- PCR:
-
Polymerase chain reaction
- SCFA:
-
Short-chain fatty acids
References
Tito RY, Chaffron S, Caenepeel C, Lima-Mendez G, Wang J, Vieira-Silva S, et al. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut. 2019;68:1180–9. https://doi.org/10.1136/gutjnl-2018-316106.
Andersen LO, Bonde I, Nielsen HB, Stensvold CR. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol Ecol. 2015. https://doi.org/10.1093/femsec/fiv072.
Rostami A, Riahi SM, Haghighi A, Saber V, Armon B, Seyyedtabaei SJ. The role of Blastocystis sp. and Dientamoeba fragilis in irritable bowel syndrome: a systematic review and meta-analysis. Parasitol Res. 2017;116:2361–71. https://doi.org/10.1007/s00436-017-5535-6.
Clark CG, van der Giezen M, Alfellani MA, Stensvold CR. Recent developments in Blastocystis research. Adv Parasitol. 2013;82:1–32. https://doi.org/10.1016/B978-0-12-407706-5.00001-0.
Cinek O, Polackova K, Odeh R, Alassaf A, Kramna L, Ibekwe MU, et al. Blastocystis in the faeces of children from six distant countries: prevalence, quantity, subtypes and the relation to the gut bacteriome. Parasit Vectors. 2021;14:399. https://doi.org/10.1186/s13071-021-04859-3.
Poulsen CS, Efunshile AM, Nelson JA, Stensvold CR. Epidemiological aspects of blastocystis colonization in children in Ilero. Nigeria Am J Trop Med Hyg. 2016;95:175–9. https://doi.org/10.4269/ajtmh.16-0074.
Mohammad NA, Al-Mekhlafi HM, Moktar N, Anuar TS. Prevalence and risk factors of Blastocystis infection among underprivileged communities in rural Malaysia. Asian Pac J Trop Med. 2017;10:491–7. https://doi.org/10.1016/j.apjtm.2017.05.001.
Oliveira-Arbex AP, David EB, Guimaraes S. Blastocystis genetic diversity among children of low-income daycare center in Southeastern Brazil. Infect Genet Evol. 2018;57:59–63. https://doi.org/10.1016/j.meegid.2017.11.005.
Wawrzyniak I, Poirier P, Viscogliosi E, Dionigia M, Texier C, Delbac F, et al. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Ther Adv Infect Dis. 2013;1:167–78. https://doi.org/10.1177/2049936113504754.
Stensvold CR, Clark CG. Current status of Blastocystis: a personal view. Parasitol Int. 2016;65:763–71. https://doi.org/10.1016/j.parint.2016.05.015.
Bart A, Wentink-Bonnema EM, Gilis H, Verhaar N, Wassenaar CJ, van Vugt M, et al. Diagnosis and subtype analysis of Blastocystis sp. in 442 patients in a hospital setting in the Netherlands. BMC Infect Dis. 2013;13:389. https://doi.org/10.1186/1471-2334-13-389.
El Safadi D, Cian A, Nourrisson C, Pereira B, Morelle C, Bastien P, et al. Prevalence, risk factors for infection and subtype distribution of the intestinal parasite Blastocystis sp. from a large-scale multi-center study in France. BMC Infect Dis. 2016;16:451. https://doi.org/10.1186/s12879-016-1776-8.
Scanlan PD, Knight R, Song SJ, Ackermann G, Cotter PD. Prevalence and genetic diversity of Blastocystis in family units living in the United States. Infect Genet Evol. 2016;45:95–7. https://doi.org/10.1016/j.meegid.2016.08.018.
Scanlan PD, Stensvold CR, Rajilic-Stojanovic M, Heilig HG, De Vos WM, O’Toole PW, et al. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol. 2014;90:326–30. https://doi.org/10.1111/1574-6941.12396.
Lhotska Z, Jirku M, Hlozkova O, Brozova K, Jirsova D, Stensvold CR, et al. A study on the prevalence and subtype diversity of the intestinal protist Blastocystis sp. in a gut-healthy human population in the Czech Republic. Front Cell Infect Microbiol. 2020;10:544335. https://doi.org/10.3389/fcimb.2020.544335.
Maloney JG, Molokin A, da Cunha MJR, Cury MC, Santin M. Blastocystis subtype distribution in domestic and captive wild bird species from Brazil using next generation amplicon sequencing. Parasite Epidemiol Control. 2020;9:e00138. https://doi.org/10.1016/j.parepi.2020.e00138S2405-6731(20)30007-6[pii]e00138[pii].
Stensvold CR, Clark CG. Pre-empting Pandora’s box: Blastocystis subtypes revisited. Trends Parasitol. 2020;36:229–32. https://doi.org/10.1016/j.pt.2019.12.009.
Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro-Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–8. https://doi.org/10.1016/j.actatropica.2012.12.011.
Stark D, Barratt J, Chan D, Ellis JT. Dientamoeba fragilis, the neglected trichomonad of the human bowel. Clin Microbiol Rev. 2016;29:553–80. https://doi.org/10.1128/CMR.00076-15.
Jokelainen P, Hebbelstrup Jensen B, Andreassen BU, Petersen AM, Roser D, Krogfelt KA, et al. Dientamoeba fragilis, a commensal in children in Danish day care centers. J Clin Microbiol. 2017;55:1707–13. https://doi.org/10.1128/JCM.00037-17.
Jirku M, Kasparova A, Lhotska Z, Obornik M, Brozova K, Petrzelkova KJ, et al. A cross-sectional study on the occurrence of the intestinal protist, Dientamoeba fragilis, in the gut-healthy volunteers and their animals. Int J Mol Sci. 2022. https://doi.org/10.3390/ijms232315407.
Stensvold CR, Arendrup MC, Molbak K, Nielsen HV. The prevalence of Dientamoeba fragilis in patients with suspected enteroparasitic disease in a metropolitan area in Denmark. Clin Microbiol Infect. 2007;13:839–42. https://doi.org/10.1111/j.1469-0691.2007.01760.x.
Barratt JL, Harkness J, Marriott D, Ellis JT, Stark D. A review of Dientamoeba fragilis carriage in humans: several reasons why this organism should be considered in the diagnosis of gastrointestinal illness. Gut Microbes. 2011;2:3–12. https://doi.org/10.4161/gmic.2.1.14755.
Windsor JJ, Johnson EH. Dientamoeba fragilis: the unflagellated human flagellate. Br J Biomed Sci. 1999;56:293–306.
Oliveira-Arbex AP, David EB, Caccio SM, Fonseca C, Martin JG, Kurokawa CS, et al. Prevalence and genetic characterization of Dientamoeba fragilis in asymptomatic children attending daycare centers. Rev Inst Med Trop Sao Paulo. 2021;63:e39. https://doi.org/10.1590/S1678-9946202163039.
Ogren J, Van Nguyen S, Nguyen MK, Dimberg J, Matussek A. Prevalence of Dientamoeba fragilis, Giardia duodenalis, Entamoeba histolytica/dispar, and Cryptosporidium in Da Nang, Vietnam, detected by a multiplex real-time PCR. APMIS. 2016;124:529–33. https://doi.org/10.1111/apm.12535.
Barry MA, Weatherhead JE, Hotez PJ, Woc-Colburn L. Childhood parasitic infections endemic to the United States. Pediatr Clin North Am. 2013;60:471–85. https://doi.org/10.1016/j.pcl.2012.12.011.
Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169:345–50.
Kemppainen KM, Lynch KF, Liu E, Lonnrot M, Simell V, Briese T, et al. Factors that increase risk of celiac disease autoimmunity after a gastrointestinal infection in early life. Clin Gastroenterol Hepatol. 2017;15:694-702 e5. https://doi.org/10.1016/j.cgh.2016.10.033.
Sanchez E, Donat E, Ribes-Koninckx C, Fernandez-Murga ML, Sanz Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl Environ Microbiol. 2013;79:5472–9. https://doi.org/10.1128/AEM.00869-13.
Kondrashova A, Mustalahti K, Kaukinen K, Viskari H, Volodicheva V, Haapala AM, et al. Lower economic status and inferior hygienic environment may protect against celiac disease. Ann Med. 2008;40:223–31. https://doi.org/10.1080/07853890701678689.
Oikarinen M, Puustinen L, Lehtonen J, Hakola L, Simell S, Toppari J, et al. Enterovirus infections are associated with the development of celiac disease in a birth cohort study. Front Immunol. 2020;11:604529. https://doi.org/10.3389/fimmu.2020.604529.
Tapia G, Chuda K, Kahrs CR, Stene LC, Kramna L, Marild K, et al. Parechovirus infection in early childhood and association with subsequent celiac disease. Am J Gastroenterol. 2021;116:788–95. https://doi.org/10.14309/ajg.0000000000001003.
Hakansson A, Andren Aronsson C, Brundin C, Oscarsson E, Molin G, Agardh D. Effects of Lactobacillus plantarum and Lactobacillus paracasei on the peripheral immune response in children with celiac disease autoimmunity: a randomized, double-blind placebo-controlled clinical trial. Nutrients. 2019. https://doi.org/10.3390/nu11081925.
Oscarsson E, Hakansson A, Andren Aronsson C, Molin G, Agardh D. Effects of probiotic bacteria Lactobacillaceae on the gut microbiota in children with celiac disease autoimmunity: a placebo-controlled and randomized clinical trial. Front Nutr. 2021;8:680771. https://doi.org/10.3389/fnut.2021.680771.
Stensvold CR, Ahmed UN, Andersen LO, Nielsen HV. Development and evaluation of a genus-specific, probe-based, internal-process-controlled real-time PCR assay for sensitive and specific detection of Blastocystis spp. J Clin Microbiol. 2012;50:1847–51. https://doi.org/10.1128/JCM.00007-12.
Sloufova M, Lhotska Z, Jirku M, Petrzelkova KJ, Stensvold CR, Cinek O, et al. Comparison of molecular diagnostic approaches for the detection and differentiation of the intestinal protist Blastocystis sp in humans. Parasite. 2022;29:30. https://doi.org/10.1051/parasite/2022029.
Maloney JG, Molokin A, Santin M. Next generation amplicon sequencing improves detection of Blastocystis mixed subtype infections. Infect Genet Evol. 2019;73:119–25. https://doi.org/10.1016/j.meegid.2019.04.013.
Santin M, Gomez-Munoz MT, Solano-Aguilar G, Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol Res. 2011;109:205–12. https://doi.org/10.1007/s00436-010-2244-9.
Verweij JJ, Mulder B, Poell B, van Middelkoop D, Brienen EA, van Lieshout L. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol Cell Probes. 2007;21:400–4. https://doi.org/10.1016/j.mcp.2007.05.006.
Hurych J, Vodolanova L, Vejmelka J, Drevinek P, Kohout P, Cinek O, et al. Freezing of faeces dramatically decreases the viability of Blastocystis sp. and Dientamoeba fragilis. Eur J Gastroenterol Hepatol. 2022;34:242–3. https://doi.org/10.1097/MEG.0000000000002327.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. https://doi.org/10.1038/nmeth.3869.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590-6. https://doi.org/10.1093/nar/gks1219.
Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, et al: vegan: Community Ecology Package. R package version 2.5–6. 2019.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. https://doi.org/10.1371/journal.pone.0061217.
Team RC. R: a language and environment for statistical computing. 3.4.2. Vienna: R Foundation for Statistical Computing; 2017.
Bach JF. Protective role of infections and vaccinations on autoimmune diseases. J Autoimmun. 2001;16:347–53. https://doi.org/10.1006/jaut.2000.0478.
Soleimani Jevinani S, Mohammad Rahimi H, Asri N, Rostami-Nejad M, Ahmadipour S, Mirjalali H. Molecular epidemiology and subtyping of Blastocystis sp. and its subtypes in celiac patients; a case control study. Microb Pathog. 2023;179:106086. https://doi.org/10.1016/j.micpath.2023.106086.
El Safadi D, Gaayeb L, Meloni D, Cian A, Poirier P, Wawrzyniak I, et al. Children of Senegal river basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect Dis. 2014;14:164. https://doi.org/10.1186/1471-2334-14-164.
Roser D, Simonsen J, Nielsen HV, Stensvold CR, Molbak K. Dientamoeba fragilis in Denmark: epidemiological experience derived from four years of routine real-time PCR. Eur J Clin Microbiol Infect Dis. 2013;32:1303–10. https://doi.org/10.1007/s10096-013-1880-2.
Ogren J, Dienus O, Lofgren S, Einemo IM, Iveroth P, Matussek A. Dientamoeba fragilis prevalence coincides with gastrointestinal symptoms in children less than 11 years old in Sweden. Eur J Clin Microbiol Infect Dis. 2015;34:1995–8. https://doi.org/10.1007/s10096-015-2442-6.
van Bruijnesteijn CLE, Dullaert-de BM, Ruijs GJ, van der Reijden WA, van der Zanden AG, Weel JF, et al. Case-control comparison of bacterial and protozoan microorganisms associated with gastroenteritis: application of molecular detection. Clin Microbiol Infect. 2015;21:592-e9-19.
de Wit MA, Koopmans MP, Kortbeek LM, van Leeuwen NJ, Vinje J, van Duynhoven YT. Etiology of gastroenteritis in sentinel general practices in the Netherlands. Clin Infect Dis. 2001;33:280–8. https://doi.org/10.1086/321875.
Dinleyici EC, Eren M, Dogan N, Reyhanioglu S, Yargic ZA, Vandenplas Y. Clinical efficacy of Saccharomyces boulardii or metronidazole in symptomatic children with Blastocystis hominis infection. Parasitol Res. 2011;108:541–5. https://doi.org/10.1007/s00436-010-2095-4.
Lepczynska M, Dzika E. The influence of probiotic bacteria and human gut microorganisms causing opportunistic infections on Blastocystis ST3. Gut Pathog. 2019;11:6. https://doi.org/10.1186/s13099-019-0287-8.
Di Biase AR, Marasco G, Ravaioli F, Dajti E, Colecchia L, Righi B, et al. Gut microbiota signatures and clinical manifestations in celiac disease children at onset: a pilot study. J Gastroenterol Hepatol. 2021;36:446–54. https://doi.org/10.1111/jgh.15183.
Bodkhe R, Shetty SA, Dhotre DP, Verma AK, Bhatia K, Mishra A, et al. Comparison of small gut and whole gut microbiota of first-degree relatives with adult celiac disease patients and controls. Front Microbiol. 2019;10:164. https://doi.org/10.3389/fmicb.2019.00164.
Quagliariello A, Aloisio I, Bozzi Cionci N, Luiselli D, D’Auria G, Martinez-Priego L, et al. Effect of Bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. 2016. https://doi.org/10.3390/nu8100660.
Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol. 2009;62:264–9. https://doi.org/10.1136/jcp.2008.061366.
Leonard MM, Valitutti F, Karathia H, Pujolassos M, Kenyon V, Fanelli B, et al. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc Natl Acad Sci USA. 2021. https://doi.org/10.1073/pnas.2020322118.
Rodrigues VF, Elias-Oliveira J, Pereira IS, Pereira JA, Barbosa SC, Machado MSG, et al. Akkermansia muciniphila and gut immune system: a good friendship that attenuates inflammatory bowel disease, obesity, and diabetes. Front Immunol. 2022;13:934695. https://doi.org/10.3389/fimmu.2022.934695.
Turjeman S, Sharon E, Levin R, Oralewska B, Szaflarska-Poplawska A, Bierla JB, et al. Celiac-the lone horse? An autoimmune condition without signals of microbiota dysbiosis. Microbiol Spectr. 2023. https://doi.org/10.1128/spectrum.01463-23.
Nieves-Ramirez ME, Partida-Rodriguez O, Laforest-Lapointe I, Reynolds LA, Brown EM, Valdez-Salazar A, et al. Asymptomatic intestinal colonization with protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems. 2018. https://doi.org/10.1128/mSystems.00007-18.
Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–76. https://doi.org/10.1099/ijs.0.02873-0.
Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–61. https://doi.org/10.1128/AEM.66.4.1654-1661.2000.
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. https://doi.org/10.1073/pnas.0804812105.
Testerman T, Li Z, Galuppo B, Graf J, Santoro N. Insights from shotgun metagenomics into bacterial species and metabolic pathways associated with NAFLD in obese youth. Hepatol Commun. 2022;6:1962–74. https://doi.org/10.1002/hep4.1944.
Bojovic K, Ignjatovic Eth I, Sokovic Bajic S, Vojnovic Milutinovic D, Tomic M, Golic N, et al. Gut microbiota dysbiosis associated with altered production of short chain fatty acids in children with neurodevelopmental disorders. Front Cell Infect Microbiol. 2020;10:223. https://doi.org/10.3389/fcimb.2020.00223.
Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27:321–32. https://doi.org/10.1038/s41591-020-01183-8.
Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–22. https://doi.org/10.1038/s41579-018-0029-9.
Zierdt CH. Blastocystis hominis–past and future. Clin Microbiol Rev. 1991;4:61–79. https://doi.org/10.1128/CMR.4.1.61.
Acknowledgements
Zuzana Lhotská from the Institute of Parasitology, Biology Centre, Czech Academy of Sciences, and Zdeněk Verner, from Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University in Vestec (BIOCEV), are sincerely thanked for preparing cultures of Blastocystis and Entamoeba. Kateřina Chudá from the Department of Paediatrics, 2nd Faculty of Medicine, Charles University, is sincerely thanked for designing the primers for Entamoeba. Anna Kjelström from the Department of Food Technology Engineering and Nutrition, Lund University, is sincerely thanked for her expert technical assistance.
Funding
Open access funding provided by Lund University. This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 874864 HEDIMED. This publication reflects only the authors' views, and the European Commission is not responsible for any use that may be made of the information it contains. The CiPP Study was funded by Stiftelsen Samariten, FoU Region Skåne, SUS fonder, Swedish Celiac Disease Foundation, Swedish Research Council, Grant/Award No.: 2018-02553, Crafoords stiftelse, Dr Per Håkanssons stiftelse, and Probi AB. JH's stay at Lund University was supported by CZ.02.2.69/0.0/0.0/18_053/0016976: International mobility of research, technical and administrative staff at Charles University; O.C. and J.H. are financed by the project National Institute of Virology and Bacteriology (Programme EXCELES, ID Project No. LX22NPO5103)—Funded by the European Union—Next Generation EU, and by COST Action CA21105, supported by COST (European Cooperation in Science and Technology).
Author information
Authors and Affiliations
Consortia
Contributions
JH performed laboratory analyses, analysed some of the data and wrote the manuscript; EO performed some laboratory analyses; AH supervised the laboratory analyses and co-lead the study; KJM and MJ contributed to the preparation of the detection assays, construction of quantitative calibrators and manuscript revision; CA designed and co-lead the study, including the subjects recruitment, metadata gathering and data management; OC designed the laboratory part of this study, analysed the data and revised the manuscript; DA conceived the CiPP study, participated in its organization, subject recruitment and data interpretation and revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Medical Faculty, Lund University, on September 8th, 2011 (Dnr 2011/335) and on September 1st, 2021 (Dnr 2021-04470). The study is registered in ClinicalTrials.gov (NCT03176095).
Consent for publication
Not applicable.
Competing interests
DA is an inventor in a patent application based on the results of the clinical trial but has signed over all legal rights to the patent to Probi AB. Probi AB has developed and supplied the study material (active and placebo products) for the trial as well as financially supported the trial with minor costs for analyzing material. None of the authors are employed by Probi AB and no salaries, consultancy fees, etc., have been paid by Probi AB to the authors in connection with the trial. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Statistical analysis—R markdown.
Additional file 2
: Figure S1. The detection primers and hydrolysis probe annealing to the consensus of several sequences of Entamoeba sp. Figure S2. Positivity of Blastocystis sp., Dieantamoeba fragilis and Entamoeba sp. in individual study samples. Figure S3. Constrained ordination (redundancy analysis) of the bacteriome community composition by Blastocystis sp. positivity, Dieantamoeba fragilis positivity and intervention with lactobacilli. Figure S4. The dispersion of samples negative for Blastocystis sp. or Dieantamoeba fragilis is significantly higher than that of their positive counterparts (P < 0.001), so testing by Permutational Multivariate Analysis of Variance would not be meaningful—its significant result may reflect not only the significant difference in centroid position but also the difference in spread. Table S1. Specific primers and probes for Entamoeba sp. quantitative PCR.
Additional file 3.
Table—Parasites detection.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hurych, J., Oscarsson, E., Håkanson, Å. et al. Effects of Lactiplantibacillus plantarum and Lacticaseibacillus paracasei supplementation on the single-cell fecal parasitome in children with celiac disease autoimmunity: a randomized, double-blind placebo-controlled clinical trial. Parasites Vectors 16, 411 (2023). https://doi.org/10.1186/s13071-023-06027-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-06027-1