Abstract
Background
Visceral leishmaniasis (VL), a life-threatening neglected tropical disease, is targeted for elimination from Nepal by the year 2026. The national VL elimination program is still confronted with many challenges including the increasingly widespread distribution of the disease over the country, local resurgence and the questionable efficacy of the key vector control activities. In this study, we assessed the status and risk of Leishmania donovani transmission based on entomological indicators including seasonality, natural Leishmania infection rate and feeding behavior of vector sand flies, Phlebotomus argentipes, in three districts that had received disease control interventions in the past several years in the context of the disease elimination effort.
Methods
We selected two epidemiologically contrasting settings in each survey district, one village with and one without reported VL cases in recent years. Adult sand flies were collected using CDC light traps and mouth aspirators in each village for 12 consecutive months from July 2017 to June 2018. Leishmania infection was assessed in gravid sand flies targeting the small-subunit ribosomal RNA gene of the parasite (SSU-rRNA) and further sequenced for species identification. A segment (~ 350 bp) of the vertebrate cytochrome b (cytb) gene was amplified from blood-fed P. argentipes from dwellings shared by both humans and cattle and sequenced to identify the preferred host.
Results
Vector abundance varied among districts and village types and peaks were observed in June, July and September to November. The estimated Leishmania infection rate in vector sand flies was 2.2% (1.1%–3.7% at 95% credible interval) and 0.6% (0.2%–1.3% at 95% credible interval) in VL and non-VL villages respectively. The common source of blood meal was humans in both VL (52.7%) and non-VL (74.2%) villages followed by cattle.
Conclusions
Our findings highlight the risk of ongoing L. donovani transmission not only in villages with VL cases but also in villages not reporting the presence of the disease over the past several years within the districts having disease elimination efforts, emphasize the remaining threats of VL re-emergence and inform the national program for critical evaluation of disease elimination strategies in Nepal.
Graphical Abstract

Similar content being viewed by others
Background
Visceral leishmaniasis (VL), also known as kala-azar in the Indian subcontinent (ISC), is the most life-threatening form of leishmaniasis, with an estimated 50,000 to 90,000 new cases occurring worldwide every year [1]. Although India, Nepal and Bangladesh used to carry two-thirds of the global VL burden in 2004, this fraction was reduced to only 18% in 2020 [2] after a regional elimination initiative was launched in 2005. This initiative targeted the elimination of the disease from the ISC by 2015 [3], a deadline later extended to 2026 [4]. The elimination target was to reduce the annual incidence below one case per 10,000 population at the implementation unit level (district level in Nepal and sub-district level in India and Bangladesh) in all endemic areas, a target which is assumed to no longer form a public health concern [5]. VL was envisaged for elimination from the ISC because of some favorable factors like humans being the only known host or reservoir, Phlebotomus argentipes sand fly as the only vector transmitting the parasite Leishmania donovani, availability of chemical-based vector control tools, rapid disease diagnostic tools and complete treatment of the disease. Accordingly, the key strategies adopted in the region and in Nepal for disease elimination are early diagnosis and complete treatment, integrated vector management with main focus on indoor residual spraying (IRS) and effective disease and vector surveillance [6].
By 2013, the VL elimination target was achieved in all 12 officially endemic districts (districts where a full transmission cycle and therefore local transmission has been formally established, warranting the presence of the national VL elimination program) mostly situated in the central and eastern plain lowland areas (also called as “terai”) of Nepal. Nonetheless, 10 years after this first achievement, Nepal has still not been able to declare the elimination of VL as a public health problem. Several districts have breached the elimination target again since 2013, with two districts above the threshold in 2022. Also, the national VL elimination program added six districts in 2016 and five more in 2021 to the endemic district’s list, after the local transmission was epidemiologically and entomologically verified [2, 6, 7]. These newly endemic districts include hilly and mountainous areas from the eastern, central and western parts of the country. At present, 42 districts are listed as endemic, 30 districts as endemic doubtful (reports of at least one locally acquired VL case in the last 10 years and full cycle of transmission is yet to be verified) and only five districts as non-endemic (no locally acquired VL cases in the last 10 years) out of 77 total districts in Nepal (unpublished data).
The continued presence of sporadic VL cases even in the districts below the elimination threshold in the historical endemic areas sustains the risk of new outbreaks, local resurgence and transmission to neighboring unaffected areas. In this context, the elimination strategies need critical evaluation, realignment and reconsideration of relevance in the current phase of sustaining the elimination target and preventing re-emergence of disease transmission [5, 8]. One of the key strategies, i.e. vector control with synthetic pyrethroid-based IRS in human and animal shelters to reduce L. donovani transmission [9, 10], is vulnerable to poor performance because of logistic constraints, irregular operational practices and limited updated knowledge on vector bionomics [11,12,13]. Vector abundance and seasonality in an area are often related to the extent of vector-human contact and Leishmania transmission [14], and up-to-date information facilitates determining the appropriate spray timing for effective IRS implementation. Furthermore, even in the absence of reported VL cases, transmission can still be ongoing, as the vast majority of infections remain asymptomatic in humans, and clinical cases can be missed or can migrate elsewhere before diagnosis. Surveillance of the prevalence of Leishmania infection in vector sand flies can help to understand to what extent the parasite is still circulating in a certain area and provide crucial information for the prediction of the risk and expansion of leishmaniasis [15,16,17,18]. The role of a sand fly species as a vector is invariably associated with the intensity of host-vector contact for blood feeding; therefore, it is undoubtedly important to identify and quantify the source of blood meals in wild-caught vector sand flies to determine the risk of Leishmania transmission in the human population [19, 20].
Several studies investigated sand fly seasonality, prevalence of Leishmania in sand flies and host preferences in the early years of the elimination initiative, mostly to assess the Leishmania transmission dynamics and to check the efficacy of various vector control tools in Nepal [13, 17, 21,22,23]. As considerable changes in the epidemiological situation have been experienced in Nepal since then, updated information on these entomological indicators is sought to guide the VL elimination efforts for sustainable elimination of the disease in the country.
In this study, we aimed to assess the status and risk of L. donovani transmission in longstanding endemic districts by collecting data on seasonality, natural L. donovani infection rate and host preference of vector sand flies from two epidemiologically contrasting areas; villages with VL cases being reported in recent years and neighboring villages within the same endemic districts but without any such cases reported during the last 10 years in the eastern part of Nepal.
Methods
Study area
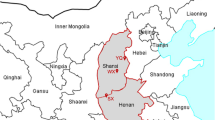
Three VL endemic districts in eastern Nepal, viz. Morang, Sunsari and Saptari (Fig. 1), were selected for field work. We selected these districts based on (i) the cumulative VL case load over the years 2014–2017 (Additional file 1: Table S1) and (ii) their proximity to the BP Koirala Institute of Health Sciences (BPKIHS), Dharan, Nepal. In each district, one village reporting VL cases in three consecutive years prior to the inception of the study was assigned as the ‘VL village’, and a second village without any reported VL cases in the last 10 years before the start of the study was selected as ‘non-VL village’. Corresponding VL and non-VL villages in each district were carefully selected within 5–10 km distance of each other to minimize the differences in spatial, ecological and housing characteristics. These VL and non-VL villages were Majhare (26.406992 N, 87.337322 E) and Pokhariya (26.401985 N, 87.363547 E) in Morang district, Amahibelha (26.466103 N, 87.188618 E) and Chimadi (26.492791 N, 87.186457 E) in Sunsari district and Belhichapena (26.524450 N, 86.677925 E) and Gamhariya Parwaha (26.541511 N, 86.663169 E) in Saptari district respectively (Fig. 1).
Map of Nepal showing visceral leishmaniasis endemicity (situation as of the year 2016–2017) (inset) and the locations of the study districts and villages. VL villages, villages reporting VL cases in three consecutive years prior to the inception of the study. Non-VL villages, villages without any reported VL cases in the last 10 years before the start of the study. The map was produced with QGIS (version 3.10.2) with open access shapefile. (https://opendatanepal.com/dataset/new-political-and-administrative-boundaries-shapefile-of-nepal)
Sand fly collection and morphological identification
In each of the selected villages, 10 households including human dwellings and mixed dwellings (cattle sheds sharing space with humans) were purposively selected for longitudinal monitoring of the sand fly density for 12 consecutive months, from July 2017 until June 2018. Centre for Disease Control and Prevention miniature light traps (Model 2836 BQ; BioQuip Products, Rancho Dominguez, CA 90220, USA) were used as a standard method of sand fly capturing. One light trap (LT) was placed in the inner lower corner of the main room or cattle shed of each of the selected households and kept functioning for 12 h starting at 18 h to 6 h. In the consecutive morning, resting sand flies were collected using mouth aspirators spending 15 min time in the same household where LT was placed. Both LT and aspirator collections were made for three consecutive nights and mornings in all 10 houses of a village every month during the study period.
We transferred the LT and aspirator collections to the BPKIHS entomology laboratory on a daily basis for the morphological identification of the sand flies using a binocular microscope and regional keys [24, 25]. All sand flies were preserved in 80% ethanol and refrigerated at 4 °C. Male P. argentipes and other species of sand flies were stored at the village level per month. Female P. argentipes were segregated and stored at the physiological stages (unfed, fed and gravid) at the household level by collection method (light trap versus aspirator) and place of collection (human dwelling and mixed dwellings) level every month.
Molecular analyses of the P. argentipes sand flies
DNA extraction
Genomic DNA was extracted from pools of gravid P. argentipes sand flies for Leishmania detection. Pooling of the gravid P. argentipes was done by collection method (LTs and aspirator), household, village and the survey month. Each pool was comprised of a minimum one to a maximum 20 sand flies. In case of more than 20 gravid sand flies collected at an event, the number was divided into two or more pools each containing not more than 20 specimens. Besides, DNA was extracted from individual specimens of blood-fed vector sand flies for blood meal source identification. For reference purpose, DNA extractions were also obtained from blood samples (approximately 5 ml each) of vertebrate hosts like buffalo, cow, goat, pig, rabbit, chicken, duck and pigeon.
The sand flies were crushed using a sterile micro-pestle and homogenized before following the DNA extraction procedure using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA elution of 50 µl was collected. The quantity and purity of the DNA extract were checked with a NanoDrop2000 spectrophotometer (Thermo Scientific, DE). DNA extracts were preserved at − 20 °C until being subjected to specific polymerase chain reactions.
Detection of Leishmania infection in vector sand flies
Assessment of Leishmania infection was done on pools of all gravid P. argentipes collected throughout the year. A TaqMan probe-based real-time PCR assay targeting the 115 bp length of the small-subunit ribosomal RNA gene of Leishmania (SSU-rRNA) was conducted on DNA extracts from the pools of gravid P. argentipes. A Leish 18S Set (LE18S) (Integrated DNA Technologies, Belgium) was used in the assay with already described and tested primers: M18S-F-L (5′-CGT AGT TGA ACT GTG GGC TGT GC-3′) and M-18S-R-L (5′-ACT CCC GTG TTT CTT GTT TCT TTG AA-3′) [17, 26], along with a double quenched L-probe (FAM-CTGGTCGTCCCGTCCATGTCGGATT-BHQ1 ZEN). The PCR reaction was carried out in a final volume of 25 µl containing 0.4 µM M18S-F-L, 0.4 µM M-18S-R-L and 0.1 µM L-probe, 1X GoTaq Probe qPCR Master Mix (Promega, USA), 0.1 mg/ml bovine serum albumin (Promega, USA) and 2.5 µl sample DNA. The PCR amplification was conducted in the Rotor-GeneQ real-time PCR System (Qiagen, Hilden, Germany). The thermal profile optimized for the reaction was the activation of the Taq polymerase present in Go Taq Probe Master Mix at 94 °C for 15 min and then 45 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 45 s followed by the final extension at 72 °C for 5 min. In each PCR run, one positive control consisting of 1 pg of L. donovani DNA (BPK282/0) and one negative control containing PCR grade water were included for quality control of the test. Additional qPCR assay targeting the conserved and highly expressed spliced-leader [27] mini-exon sequence [28] was performed to verify the outcomes of the SSU-rRNA qPCR assay.
After the detection of the Leishmania-positive pools, Leishmania species identification was performed by amplification and sequencing of part of the internal transcribed spacer 1 (ITS-1) region [29, 30]. SSU-rRNA and SL-RNA positive samples were subjected to an ITS-1 nested PCR. A 25 µl PCR reaction mixture contained 1× GoTaq® G2 Master Mix (Promega, USA), 5 µl sample and 0.5 µM forward (5ʹ-CTG GAT CAT TTT CCG ATG-3ʹ) and reverse (5ʹ-TGA TAC CAC TTA TCG CAC TT-3ʹ) primer. Samples were tested on a Bio-Rad T100 thermal cycler using a 5 min activation step at 95 °C, 35 amplification cycles under optimized conditions (30 s at 95 °C, 30 s at 53 °C and 30 s at 72 °C) and a 10 min final elongation at 72 °C. PCR products were checked by 1% agarose gel electrophoresis and purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) after which they were subjected to a nested PCR reaction using the same conditions as described above. PCR samples were again checked by 1% agarose gel electrophoresis and purified with a QIAquick PCR Purification Kit after which they were sent for Sanger sequencing on an Applied Biosystems 3730XL DNA Analyzer at the Neuromics Support Facility of the University of Antwerp. Leishmania species identification was analyzed using SnapGene and NCBI BLAST.
Detection of blood meal sources
A targeted ~ 350 bp segment of vertebrate cytochrome b (Cytb) gene was amplified using the universal primers Cytb1-F (L14841: 5′-CCA TCC AAC ATC TCA GCA TGA TGA AA-3′) and Cytb2-R (H15149: 5′-GCC CCT CAG AAT GAT ATT TGT CCT CA-3′) [31, 32] for host identification. Each PCR was performed on a 25 μl reaction mixture containing 1× GoTaq Green Master Mix with 2 mM MgCl2 (Promega, USA), 0.4 μM each of two primers (Biolegio, The Netherlands) and 5 μl DNA template. Each set of PCR reactions was accompanied by a negative control having PCR grade water and a positive control having DNA extract from human blood. The PCR reaction was performed on an Eppendorf Mastercycler pro PCR System (Hamburg, Germany) setting the thermal profile as pre-activation step at 95 °C for 2 min, followed by 40 cycles of denaturation at 92 °C for 30 s, annealing at 55 °C for 45 s and extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. The PCR products, positive and negative controls were loaded on 2% agarose gel (Eurogentec, Belgium), stained with ethidium bromide (Promega, USA) and examined under a UV trans-illuminator/gel documentation system (Syngene, Cambridge, UK). PCR products (amplicons) were outsourced for Sanger sequencing at the Macrogen Company, South Korea. All generated sequences were checked and edited in BioEdit Sequence Alignment Editor 7.0.5.3 [33] and queried against NCBI BLAST.
Molecular validation of P. argentipes identification
A subsample of female P. argentipes subjected to Leishmania infection assessment (n = 120) and blood meal analysis (n = 31) was verified by DNA barcoding method based on the amplification of targeted 650 bp fragment length of mitochondrial cytochrome c oxidase subunit I (COI) gene as described by Folmer et al., Hebert et al. and Kumar et al. [34,35,36].
Data management and statistical analysis
Entomological data were recorded in a pre-tested paper-based data collection tool and entered in a database developed in Epi Info version 3.5.1 [37]. All statistical analyses and graphical plotting were performed with R software version 4.1.0 [38].
Sand fly abundance and seasonality analysis
Sand fly abundance and seasonality analysis were based on the LT collections only, considering them as the standard method of vector collection. Sand flies captured by mouth aspirators were excluded from this analysis to avoid any possible collection bias. Mean number of P. argentipes sand flies captured per trap-night per house is represented in a line graph by month, type of village and district. We fitted a generalized linear model (GLM) with a negative binomial distribution to assess the sand fly abundance in function of the type of village, district and month. We used a Bayesian approach to better account for the uncertainty of the estimates using the R package “rstanarm” [39, 40]. This approach produces a 95% credible interval, which means that given the observations, there is a 95% chance that the value lies between the limits of the credible interval [41]. Results of the analysis are represented as an incidence rate ratio (IRR) and credible interval (CI) at 95%.
Estimation of the prevalence of Leishmania infection rate
For the assessment of Leishmania infection, all gravid P. argentipes sand flies collected in both LTs and aspirators were used. The prevalence of Leishmania infection in pools of unequal size was estimated by a Bayesian Markov Chain Monte Carlo (MCMC) model as described by Speybroeck et al. [42, 43]. In this method, prior knowledge of the diagnostic test (in this case, the SSU-rRNA PCR method for detection of Leishmania in pools of vector sand flies) characteristics (sensitivity and specificity) was incorporated with the test results. We assumed an imperfect sensitivity, ranging uniformly between 60 and 95%, and a perfect specificity for the PCR test [26]. The result obtained was a posterior probability distribution of the prevalence; the mean and credible intervals at 95% were then calculated from it. Prevalence was estimated using the “truePrevPools” function in the R package “prevalence” [44], and credible intervals (CI) were calculated in the R package “bayestestR” [45, 46].
Blood meal analysis
A subsample of 50 blood-fed P. argentipes collected in both light traps and aspirators from mixed dwellings of each village during the initial months of the surveys was used for blood meal analysis. Host preference was calculated as the percentage of P. argentipes sand flies fed on a specific host relative to the total number of blood-fed P. argentipes tested per village category (i.e. VL versus non-VL villages).
Results
Sand fly abundance and seasonality
A total of 39,692 Phlebotomine sand flies were captured by both CDC light traps and manual aspirators from six villages of three districts during the 12 months of the survey. The vector species P. argentipes (n = 31,382; 79.1%) was dominant compared to other sand fly species Phlebotomus papatasi (n = 1632; 4.1%) and Sergentomyia spp. (n = 6678; 16.8%).
Considering the vector sand flies collected in light traps only (n = 20,101), males (n = 11,725; 58.3%) outnumbered females (n = 8376; 41.7%). About double the number of P. argentipes sand flies were captured per trap-night per house in non-VL villages (n = 12.66; IRR = 1.90; CI at 95% = 1.66–2.17) compared to VL villages (n = 6.13). Distinct variation in vector density was observed at the district level with higher per trap-night per house collections in Saptari (n = 15.47; IRR = 1.05; CI at 95% = 0.89–1.23) and lower in Sunsari (n = 3.35; IRR = 0.36; CI at 95% = 0.31–0.41) compared to collections from Morang (n = 9.35). At district level, vector density per trap-night per house was lower in the non-VL village (n = 5.55; IRR = 0.53, CI at 95% = 0.44–0.63) than in the VL village (n = 13.12) in Morang, higher in the non-VL village (n = 3.79; IRR = 1.53, CI at 95% = 1.28–1.85) than in the VL village (n = 2.92) in Sunsari and almost 12 times higher in the non-VL village (n = 28.69; IRR = 12.7, CI at 95% = 10.31–15.76) than in the VL village (n = 2.41) in Saptari (Table 1).
Month-wise abundance and seasonality of vector sand flies varied at different degrees in all three districts (Table 1, Fig. 2). In Morang, peaks in sand fly density were seen in July and September 2017 in the VL village and in July 2017 and June 2018 in the non-VL village (Table 1, Fig. 2). In Sunsari, peaks in sand fly density were observed during November 2017 and June 2018 in the VL village, while density peaks were not very clear in the non-VL village. In Saptari, many P. argentipes were captured in the non-VL village compared to all other villages year round, and the highest density peaks were observed in the months of July and November in 2017 and June in 2018. In the VL village of the same district, density peaks were seen in the months of March and May 2018. Finally, in the analysis based on the cumulative collections from all the villages, peaks in P. argentipes density were marked in the months of June and July and from September to November while the density dropped to almost zero in the months of January and February (Fig. 2, Additional file 2: Table S2).
Prevalence of Leishmania infection in vector sand flies
Leishmania infection was assessed in 796 pools of 3735 gravid P. argentipes sand flies in total. Leishmania infection was detected in four (1.37%) pools out of 293 from VL villages and one (0.2%) pool out of 503 from non-VL villages. The estimated prevalence of Leishmania infection in vector sand flies was 2.2% (CI at 95% = 1.1%–3.7%) in VL villages and 0.6% (CI at 95% = 0.2%–1.3%) in non-VL villages (Table 2). These Leishmania-infected vector sand flies were collected in the months of May, September and November. Three of five SSU-rRNA-positive pools were confirmed using the SL-RNA PCR. The Leishmania species was identified and confirmed as L. donovani in two pools from VL villages and one pool from a non-VL village. ITS-1 nested PCR was unsuccessful in two positive pools from a VL village (Table 3).
Blood meal sources and host preference patterns
A sample of 301 of 3443 fed P. argentipes collected from mixed dwellings in all six villages was assessed for identification of the source of blood meal, and vertebrate host species were successfully identified in 295. We submitted selected sequences of the cytb gene of the host species to GenBank (accession nos. OQ535519–OQ535565). The most common sources of the blood meal were humans (63.4%; 187/295), followed by domestic buffaloes (18.9%; 56/295), cows (13.9%; 41/295), goats (3.1%; 9/295) and pigs (0.7%; 2/295). In both categories of villages, P. argentipes fed primarily on humans (52.7%; 78/148 in VL vs. 74.2%; 109/147 in non-VL villages). Buffaloes (31.8%; 47/148) in VL villages and cows (16.3%; 24/147) in non-VL villages were the second preferred hosts over goats (2.7%; 4/148 in VL vs. 3.4%, 5/147 in non-VL villages). A blood meal from pigs was present in only two fed P. argentipes from VL villages (Fig. 3).
Molecular validation of P. argentipes species
The samples of morphologically identified P. argentipes sand flies subjected to the assessment of L. donovani infection and blood meal analysis were confirmed as the same species by DNA barcoding method.
Discussion
Phlebotomus argentipes sand fly density varied among districts and village types. In one of the districts, P. argentipes density was higher in the VL village than in the non-VL village, the other districts had higher collections in non-VL villages. The P. argentipes density fluctuated with the season with an increase in June and July and again in September to November. Interestingly, L. donovani-infected vector sand flies were present in both VL and non-VL villages. Furthermore, P. argentipes showed a high affinity for human blood in mixed dwellings in both types of villages.
Information on seasonality of vector sand flies is crucial to identify the peak season of human-vector contact and thus the period of maximum risk of L. donovani transmission. Based on the previous studies and information on seasonality [13, 47, 48], the national VL elimination program in Nepal has been following the schedule of two rounds of IRS using synthetic pyrethroids (alphacypermethrin, deltamethrin or lambda-cyhalothrin) in the reported VL areas or villages, the first round in the months of May–June and second in September–October [6]. The observed seasonality of vector sand fly density in this study was similar to other studies conducted in Nepal [13, 47] and bordering areas of India [49,50,51] and aligns with the existing practice of IRS. Nevertheless, the seasonality of vector abundance is conducive for Leishmania transmission throughout the year except in the months of January and February and raises concerns about the suitability of two rounds of IRS with pyrethroids, the insecticide group that is reported effective to reduce the vector sand fly density up to 4 weeks only in Nepal and Bangladesh [12, 27, 52, 53]. These findings argue for the re-evaluation of vector control strategy in terms of the number of IRS rounds to be considered to cover the long transmission season or to switch or adapt to alternative insecticides that have proven longer residual impact on the sand fly density.
The Leishmania infection in vector sand flies as another key indicator of ongoing transmission was also assessed in this study. Some of the previous studies conducted in Nepal and neighboring parts of India during the earlier years of VL elimination showed a range of 0.5% to 17.4% prevalence of Leishmania infection in vector sand flies [15,16,17, 54,55,56]. In these investigations, prevalence of infection, however, was supposedly influenced by the type of PCR test used, the statistical method to estimate infection rate and the physiological stages of the female P. argentipes (unfed, fed and gravid). Thus, the selection of only gravid P. argentipes for the Leishmania infection assessment in this study was to ensure the sand flies had taken a blood meal at least once and that the meal had already been digested. This way, we assumed the sand flies positive for Leishmania to be a proxy for infective sand flies and thus able to transmit the parasite during their next blood meal. Two PCR techniques were used for quality assurance and molecular identification confirmed the L. donovani species in the infected vector sand flies. All four L. donovani-infected pools from VL villages originated from either the house of an ex-VL patient or directly adjoining houses of the same. Interestingly, we detected an L. donovani-positive pool from a non-VL village as well, supporting the findings of the existence of asymptomatic infection in the human population in the areas with zero reported VL cases for more than a decade prior to the study [57]. The presence of infected vector sand flies even in the so-called non-VL village indicates the circulating Leishmania parasite in vector and human populations and could possibly cause resurgence of cases in these areas if left unnoticed or neglected. Thus, it is anticipated the program needs to remain vigilant for the existence of transmission even at times when VL case reporting from the longstanding endemic districts is scanty or null.
We found that vector sand flies collected from mixed dwellings had a high affinity towards humans. In mixed dwellings, sand flies were assumed to have equal access to the various types of hosts and minimal influence of biotopes on the feeding behavior. The high human blood index reported in previous studies from Nepal [21, 22] well supports the findings of our study. Despite P. argentipes being marked as an opportunistic feeder, many of the recent studies reported a high human blood index in collections from various biotopes including mixed dwellings [19, 50]. From our observations, we do not have evidence of a substantial change in feeding behavior (i.e. anthropophilic to zoophilic) of P. argentipes even after years of IRS in endemic districts; hence, the species still possesses a high potential for parasite transmission in the domestic environment.
The main strength of this study is that we assessed the risk of ongoing L. donovani transmission in VL endemic districts during the elimination era based on multiple entomological indicators. These indicators were well documented from two contrasting VL epidemiological settings, i.e. VL and VL non-villages within endemic districts, which, to our best knowledge, is the first study of this kind in Nepal. This study is also the first in 15 years to update our knowledge on seasonality, prevalence of Leishmania infection and host preference of the vector sand flies in endemic districts of Nepal. However, this entomological evidence would have been complemented by epidemiological and serological data for a complete situation analysis of L. donovani transmission in these VL endemic areas where the caseload has been reduced below the elimination threshold in recent years.
Conclusions
Our findings highlight the risk of ongoing L. donovani transmission in villages with and without reported VL cases within VL endemic districts included in the disease elimination program and that P. argentipes is still an efficient vector. The favorable entomological indicators for Leishmania transmission in non-VL villages emphasize the remaining threats to the VL elimination program and show that it needs to remain vigilant for the re-appearance of transmission even years after the last VL case has been reported from the endemic districts. In addition, we recommend that the program should strengthen the entomological and epidemiological Leishmania surveillance in Nepal to better adapt the disease elimination strategies. Also, we recommend conducting similar studies in other VL endemic districts representing various geo-ecological zones and administrative regions to monitor Leishmania transmission and track the progress towards elimination.
Availability of data and materials
All datasets generated and analyzed during the study are included in the manuscript. The generated sequences for blood meal identification are submitted in GenBank under accession nos. OQ535519–OQ535565.
References
World Health Organization. Leishmaniasis; 2023. https://www.who.int/news-room/fact-sheets/detail/leishmaniasis.
Ruiz-Postigo JA, Jain S, Mikhailov A, Maia-Elkhoury AN, Valadas S, Warusavithana S, et al. Global leishmaniasis surveillance: 2019–2020, a baseline for the 2030 roadmap. Weekly epidemiological record. WHO; 2021. 3 September Report No.: 35.
World Health Organization. Accelerating work to overcome the global impact of neglected tropical diseases. A roadmap for implementation: World Health Organization; 2012.
World Health Organization. Regional strategic framework for accelerating and sustaining elimination of kala-azar in the South-East Asia Region: 2022–2026. World Health Organization, Regional Office for South-East Asia. New Delhi: World Health Organization; 2022.
Olliaro PL, Shamsuzzaman TAKM, Marasini B, Dhariwal AC, Be-Nazir A, Mondal D, et al. Investments in research and surveillance are needed to go beyond elimination and stop transmission of Leishmania in the Indian subcontinent. PLoS Negl Trop Dis. 2017;11:e0005190. https://doi.org/10.1371/journal.pntd.0005190.
Epidemiology and disease control division. National guideline on kala-azar elimination program (Updated) 2019: Department of health services, ministry of health and population, government of Nepal, Teku, Kathmandu; 2019.
Pandey K, Dumre SP, Shah Y, Acharya BK, Khanal L, Pyakurel UR, et al. Forty years (1980–2019) of visceral leishmaniasis in Nepal: trends and elimination challenges. Trans R Soc Trop Med Hyg. 2023;117:460–9. https://doi.org/10.1093/trstmh/trad001.
Rijal S, Uranw S, Chappuis F, Picado A, Khanal B, Paudel IS, et al. Epidemiology of Leishmania donovani infection in high-transmission foci in Nepal. Trop Med Int Health. 2010;15:21–8. https://doi.org/10.1111/j.1365-3156.2010.02518.x.
Das ML, Roy L, Rijal S, Paudel IS, Picado A, Kroeger A, et al. Comparative study of kala-azar vector control measures in eastern Nepal. Acta Trop. 2010;113:162–6. https://doi.org/10.1016/j.actatropica.2009.10.012.
Joshi AB, Das ML, Akhter S, Chowdhury R, Mondal D, Kumar V, et al. Chemical and environmental vector control as a contribution to the elimination of visceral leishmaniasis on the Indian subcontinent: cluster randomized controlled trials in Bangladesh, India and Nepal. BMC Med. 2009;7:54. https://doi.org/10.1186/1741-7015-7-54.
Cameron MM, Acosta-Serrano A, Bern C, Boelaert M, den Boer M, Burza S, et al. Understanding the transmission dynamics of Leishmania donovani to provide robust evidence for interventions to eliminate visceral leishmaniasis in Bihar, India. Parasit Vectors. 2016;9:25. https://doi.org/10.1186/s13071-016-1309-8.
Huda MM, Mondal D, Kumar V, Das P, Sharma SN, Das ML, et al. Toolkit for monitoring and evaluation of indoor residual spraying for visceral leishmaniasis control in the Indian subcontinent: application and results. J Trop Med. 2011;2011:876742. https://doi.org/10.1155/2011/876742.
Picado A, Das ML, Kumar V, Dinesh DS, Rijal S, Singh SP, et al. Phlebotomus argentipes seasonal patterns in India and Nepal. J Med Entomol. 2010;47:283–6. https://doi.org/10.1603/me09175.
Ready PD. Biology of phlebotomine sand flies as vectors of disease agents. Annu Rev Entomol. 2013;58:227–50. https://doi.org/10.1146/annurev-ento-120811-153557.
Tiwary P, Kumar D, Mishra M, Singh RP, Rai M, Sundar S. Seasonal variation in the prevalence of sand flies infected with Leishmania donovani. PLoS ONE. 2013;8:e61370. https://doi.org/10.1371/journal.pone.0061370.
Tiwary P, Kumar D, Singh RP, Rai M, Sundar S. Prevalence of sand flies and Leishmania donovani infection in a natural population of female Phlebotomus argentipes in Bihar State, India. Vector Borne Zoo Dis. 2012;12:467–72. https://doi.org/10.1089/vbz.2011.0808.
Bhattarai NR, Das ML, Rijal S, van der Auwera G, Picado A, Khanal B, et al. Natural infection of Phlebotomus argentipes with Leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans R Soc Trop Med Hyg. 2009;103:1087–92. https://doi.org/10.1016/j.trstmh.2009.03.008.
Kato H, Uezato H, Gomez EA, Terayama Y, Calvopiña M, Iwata H, et al. Establishment of a mass screening method of sand fly vectors for Leishmania infection by molecular biological methods. Am J Trop Med Hyg. 2007;77:324–9.
Garlapati RB, Abbasi I, Warburg A, Poché D, Poché R. Identification of bloodmeals in wild caught blood fed Phlebotomus argentipes (Diptera: Psychodidae) using cytochrome b PCR and reverse line blotting in Bihar, India. J Med Entomol. 2012;49:515–21. https://doi.org/10.1603/me11115.
Killick-Kendrick R. The biology and control of phlebotomine sand flies. Clin Dermatol. 1999;17:279–89. https://doi.org/10.1016/S0738-081X(99)00046-2.
Burniston I, Roy L, Picado A, Das M, Rijal S, Rogers M, et al. Development of an enzyme-linked immunosorbent assay to identify host-feeding preferences of Phlebotomus species (Diptera: Psychodidae) in endemic foci of visceral leishmaniasis in Nepal. J Med Entomol. 2010;47:902–6. https://doi.org/10.1603/me09184.
Picado A, Kumar V, Das M, Burniston I, Roy L, Suman R, et al. Effect of untreated bed nets on blood-fed Phlebotomus argentipes in kala-azar endemic foci in Nepal and India. Mem Inst Oswaldo Cruz. 2009;104:1183–6. https://doi.org/10.1590/S0074-02762009000800018.
Pandey K, Pant S, Kanbara H, Shuaibu MN, Mallik AK, Pandey BD, et al. Molecular detection of Leishmania parasites from whole bodies of sandflies collected in Nepal. Parasitol Res. 2008;103:293–7. https://doi.org/10.1007/s00436-008-0967-7.
Kalra NL, Bang YH. Manuals on entomology in visceral leishmaniasis. New Delhi: World Health Organization, SEARO; 1988.
Lewis DJ. A taxonomic review of the genus Phlebotomus (Diptera: Psychodidae). Bull Br Museum Entomol Nat Hist. 1982;45:121–209.
Deborggraeve S, Boelaert M, Rijal S, De Doncker S, Dujardin JC, Herdewijn P, et al. Diagnostic accuracy of a new Leishmania PCR for clinical visceral leishmaniasis in Nepal and its role in diagnosis of disease. Trop Med Int Health. 2008;13:1378–83. https://doi.org/10.1111/j.1365-3156.2008.02154.x.
Chowdhury R, Chowdhury V, Faria S, Islam S, Maheswary NP, Akhter S, et al. Indoor residual spraying for kala-azar vector control in Bangladesh: a continuing challenge. PLoS Negl Trop Dis. 2018;12:e0006846. https://doi.org/10.1371/journal.pntd.0006846.
Eberhardt E, Van den Kerkhof M, Bulté D, Mabille D, Van Bockstal L, Monnerat S, et al. Evaluation of a pan-Leishmania Spliced-leader RNA detection method in human blood and experimentally infected syrian golden hamsters. J Mol Diagn. 2018;20:253–63. https://doi.org/10.1016/j.jmoldx.2017.12.003.
Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–58. https://doi.org/10.1016/s0732-8893(03)00093-2.
El Tai NO, Osman OF, El Fari M, Presber W, Schönian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg. 2000;94:575–9. https://doi.org/10.1016/s0035-9203(00)90093-2.
Steuber S, Abdel-Rady A, Clausen P-H. PCR-RFLP analysis: a promising technique for host species identification of blood meals from tsetse flies (Diptera: Glossinidae). Parasitol Res. 2005;97:247–54. https://doi.org/10.1007/s00436-005-1410-y.
Pareyn M, Kochora A, Van Rooy L, Eligo N, Vanden Broecke B, Girma N, et al. Feeding behavior and activity of Phlebotomus pedifer and potential reservoir hosts of Leishmania aethiopica in southwestern Ethiopia. PLoS Negl Trop Dis. 2020;14:e0007947. https://doi.org/10.1371/journal.pntd.0007947.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Asids Symp Ser. 1999;41:95–8.
Folmer O, Black M, Wr H, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial Cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–21. https://doi.org/10.1098/rspb.2002.2218.
Kumar NP, Srinivasan R, Jambulingam P. DNA barcoding for identification of sand flies (Diptera: Psychodidae) in India. Mol Ecol Resour. 2012;12:414–20. https://doi.org/10.1111/j.1755-0998.2012.03117.x.
Centre for Disease Control and Prevention. Epi Info™, a database and statistics program for public health professionals. Division of Health Informatics & Surveillance (DHIS), Center for Surveillance, Epidemiology & Laboratory Services (CSELS), Atlanta, GA, USA; 2008.
R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2021.
Goodrich B, Gabry J, Ali I, Brilleman S. rstanarm: Bayesian applied regression modeling via Stan. R package version 2.21.1; 2020.
Brilleman SL, Crowther MJ, Moreno-Betancur M, Buros Novik J, Wolfe R. Joint longitudinal and time-to-event models via Stan. StanCon 2018. 10–12 Jan 2018. Pacific Grove, CA, USA; 2018.
Gardner I. The utility of Bayes’ theorem and Bayesian inference in veterinary clinical practice and research. Aust Vet J. 2002;80:758–61. https://doi.org/10.1111/j.1751-0813.2002.tb11347.x.
Speybroeck N, Devleesschauwer B, Joseph L, Berkvens D. Misclassification errors in prevalence estimation: bayesian handling with care. Int J Public Health. 2013;58:791–5. https://doi.org/10.1007/s00038-012-0439-9.
Speybroeck N, Williams C, Lafia K, Devleesschauwer B, Berkvens D. Estimating the prevalence of infections in vector populations using pools of samples. Med Vet Entomol. 2012;26:361–71.
Devleesschauwer B, Torgerson P, Charlier J, Levecke B, Praet N, Roelandt S, et al. Prevalence: tools for prevalence assessment studies. R package version 0.4.0; 2015.
Makowski D, Ben-Shachar MS, Lüdecke D. beyestestR: describing effects and their uncertainty, existence and significance within the Bayesian framework. J Open Source Softw. 2019;4:1541. https://doi.org/10.21105/joss.01541.
Makowski D, Ben-Shachar MS, Chen SHA, Lüdecke D. Indices of effect existence and significance in the bayesian framework. Front Psychol. 2019;10:2767. https://doi.org/10.3389/fpsyg.2019.02767.
Das ML, Karki P, Koirala S, Parija SC. Entomological study of sand fly vector of kala-azar in Eastern Nepal. Health Renaissance. 2007;1:23–8.
Shrestha SL, Pant SK. Seasonal distribution of phlebotomine sand flies-vector of visceral leishmaniasis. J Nepal Med Assoc. 2003;32:237–46. https://doi.org/10.31729/jnma.1267.
Sardar AA, Chatterjee M, Jana K, Saha P, Maji AK, Guha SK, et al. Seasonal variation of sand fly populations in Kala-azar endemic areas of the Malda district, West Bengal, India. Acta Trop. 2020;204:105358. https://doi.org/10.1016/j.actatropica.2020.105358.
Poché DM, Garlapati RB, Mukherjee S, Torres-Poché Z, Hasker E, Rahman T, et al. Bionomics of Phlebotomus argentipes in villages in Bihar, India with insights into efficacy of IRS-based control measures. PLoS Negl Trop Dis. 2018;12:e0006168. https://doi.org/10.1371/journal.pntd.0006168.
Malaviya P, Hasker E, Picado A, Mishra M, Geertruyden JPV, Das ML, et al. Exposure to Phlebotomus argentipes (Diptera, Psychodidae, Phlebotominae) sand flies in rural areas of bihar, india: the role of housing conditions. PLoS ONE. 2014;9:e106771. https://doi.org/10.1371/journal.pone.0106771.
Banjara MR, Das ML, Gurung CK, Singh VK, Joshi AB, Matlashewski G, et al. Integrating case detection of visceral leishmaniasis and other febrile illness with vector control in the post-elimination phase in Nepal. Am J Trop Med Hyg. 2019;100:108–14. https://doi.org/10.4269/ajtmh.18-0307.
Chowdhury R, Huda MM, Kumar V, Das P, Joshi AB, Banjara MR, et al. The Indian and Nepalese programmes of indoor residual spraying for the elimination of visceral leishmaniasis: performance and effectiveness. Ann Trop Med Parasitol. 2011;105:31–5. https://doi.org/10.1179/136485911X12899838683124.
Basnyat S, Banjara MR, Ghimire P, Matlashewski G, Singh A. Investigation of visceral leishmaniasis transmission in selected districts of Nepal. J Nepal Health Res Counc. 2022;20:194–201. https://doi.org/10.33314/jnhrc.v20i01.4161.
Ostyn B, Uranw S, Bhattarai NR, Das ML, Rai K, Tersago K, et al. Transmission of Leishmania donovani in the hills of Eastern Nepal, an outbreak investigation in Okhaldhunga and Bhojpur Districts. PLoS Negl Trop Dis. 2015;9:e0003966. https://doi.org/10.1371/journal.pntd.0003966.
Uranw S, Hasker E, Roy L, Meheus F, Das ML, Bhattarai NR, et al. An outbreak investigation of visceral leishmaniasis among residents of Dharan town, eastern Nepal, evidence for urban transmission of Leishmania donovani. BMC Infect Dis. 2013;13:21. https://doi.org/10.1186/1471-2334-13-21.
Cloots K, Uranw S, Ostyn B, Bhattarai NR, Le Rutte E, Khanal B, et al. Impact of the visceral leishmaniasis elimination initiative on Leishmania donovani transmission in Nepal: a 10-year repeat survey. Lancet Glob Health. 2020;8:e237–43. https://doi.org/10.1016/S2214-109X(19)30536-4.
Acknowledgements
We thank all the female community health volunteers from the respective study villages for guiding us through the village and facilitating communication. We also thank the vector control officers from the respective district public health offices for helping us to select the study villages. In addition, we acknowledge the entomology technicians for their assistance in sand fly collections.
Funding
This research was supported by the doctoral fellowship (grant no. 716228/40/70, to LR) at the Institute of Tropical Medicine, Antwerp, Belgium.
Author information
Authors and Affiliations
Contributions
LR, KC, SU, MLD, EH and WVB were responsible for the study design, supervised the data collection and contributed to the writing of the manuscript. LR, KR, NRB, RH and GC performed molecular laboratory work. LR and TS did the statistical analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical clearance for the study was obtained from the Institutional Review Committee of BP Koirala Institute of Health Sciences, Dharan, Nepal (registration no. 475/073/074-IRC). Informed written consent was obtained from the household owners where sand flies were collected.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
The cumulative caseload in VL endemic and non-endemic districts in the past 3 years, 2014–16, prior to the inception of the sand fly collection.
Additional file 2: Table S2
: The total number of Phlebotomus argentipes collected by CDC light traps by district, type of village and month (30 trap-nights per village per month).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Roy, L., Cloots, K., Uranw, S. et al. The ongoing risk of Leishmania donovani transmission in eastern Nepal: an entomological investigation during the elimination era. Parasites Vectors 16, 404 (2023). https://doi.org/10.1186/s13071-023-05986-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05986-9