Abstract
Background
Malaria control relies mainlyon insecticide-based tools. However, the effectiveness of these tools is threatened by widespread insecticide resistance in malaria vectors, highlighting the need for alternative control approaches. The endosymbiont Asaia has emerged as a promising candidate for paratransgenic control of malaria, but its biology and genetics still need to be further analyzed across Africa. Here, we investigated the prevalence of Asaia and its maternal transmission in the natural population of Anopheles mosquitoes in Cameroon.
Methods
Indoor-resting adult mosquitoes belonging to four species (An. coluzzii, An. arabiensis, An. funestus and An. gambiae) were collected from eight localities across Cameroon from July 2016 to February 2020. PCR was performed on the Asaia-specific 16S ribosomal RNA gene, and samples positive by PCR for Asaia were confirmed by Sanger sequencing and phylogenetic analysis. The vertical transmission of Asaia was investigated by screening F1 mosquitoes belonging to F0 Asaia-positive females.
Results
A total of 895 mosquitoes were screened. We found 43% (384) Asaia infection prevalence in four mosquito species. Phylogenetic analysis revealed that Asaia from Cameroon clustered together with the strains of Asaia isolated from other parts of the world. In addition, seven nucleotide sequence variants were found with low genetic diversity (π = 0.00241) and nucleotide sequence variant diversity (Hd = 0.481). Asaia was vertically transmitted with high frequency (range from 42.5 to 100%).
Conclusions
This study provides field-based evidence of the presence of Asaia in Anopheles mosquitoes in Cameroon for exploitation as a symbiont in the control of malaria in sub-Saharan Africa.
Graphical abstract

Similar content being viewed by others
Background
Malaria is considered one of the most devastating diseases in sub-Saharan Africa. The World Health Organization (WHO) World malaria report 2019 recorded 216 million cases of malaria and 409,000 deaths worldwide in 2019 [1]. In Cameroon, malaria is endemic, with the whole population being at risk [2]. An estimated 23.6% of consultation in health centres, 68.7% of deaths in children under 5 years, and 16.9% of deaths in pregnant women are malaria-related cases [2, 3]. Malaria transmission in Cameroon is principally driven by species of the Anopheles gambiae complex (including An. gambiae sensu stricto [s.s.], An. arabiensis and An. coluzzii) and members of the An. funestus group (notably An. funestus s.s.) [4]. In this regard, insecticide-based vector control interventions, such as long-lasting insecticidal nets (LLINs) and indoor residual spraying (IRS) [5], have been the cornerstone of malaria prevention efforts.
However, widespread insecticide and drug resistance in mosquitoes and parasites, respectively, and the absence of an effective malaria vaccine are major obstacles weakening malaria control efforts across Africa, including Cameroon [6,7,8]. Therefore, the pursuit of novel, eco-friendly alternative tools is urgently needed to complement traditional malaria control strategies. In particular, the development of endosymbiont-based control strategies is receiving increasing attention, as microbes have desirable properties for vector control [9,10,11]. One approach, known as paratransgenesis, aims to inhibit the development of the parasites or to interfere with their competence and fitness by expressing effector molecules from symbiotic bacteria [12,13,14]. Asaia, a genus of acetic bacteria belonging to the Acetobacteraceae family [15, 16], has been identified as a promising candidate for paratransgenesis, since its characteristics support its use in vector control applications. Asaia has been isolated in malaria vectors such as An. stephensi [17], An. gambiae [18] and An. funestus [19]. It is localized in different organs of the Anopheles mosquito including the midgut, salivary glands and the reproductive system of both females and males [20]. Asaia is easy to cultivate and susceptible to genetic modification with exogenous DNA [18, 20]. It is a plant-mediated symbiont in which horizontal infection occurs through an oral route when mosquitoes feed on plant nectar [21, 22]. In addition to horizontal transmission, a venereal pattern during mating and vertical dissemination of Asaia to the next generation occurs via egg smearing [18, 23]. This highlights the symbiont’s ability to spread in mosquito populations [20]. For example, Asaia sp. SF2.1 has been demonstrated to be transmitted from female mosquitoes to their progeny through egg smearing [17]. Asaia has also been shown to facilitate the growth and reproduction of the mosquitoes seen during larval development in An. stephensi and An. gambiae [24, 25]. In addition, as shown in An. stephensi, co-localization of Asaia with a Plasmodium parasite in the host gut and the salivary gland plays a role in immune regulation through the activation of host antimicrobial peptides without self-inhibition [20].
Stable natural infection of Asaia in Anopheles mosquitoes has been shown in different species of Anopheles around the world [16, 17, 26] and more recently in African malaria vectors, notably An. gambiae, An. funestus and An. coluzzii [19, 27]. To our knowledge, only one report of Asaia has been documented in An. coluzzii and An. gambiae from Yaoundé, Cameroon, which focused on its detection by 454 pyrosequencing [28]. There is a paucity of available data on the prevalence of Asaia in other Anopheles species and in other localities across the country. In addition, the genetic diversity and the stability of the maternal transmission of Asaia in Anopheles mosquito are yet to be assessed in the Cameroonian context.
Investigating the genetic diversity and the maternal transmission of natural Asaia strains in Anopheles populations would allow a greater understanding of how this bacterium could influence malaria transmission in field populations and identify candidate strains for paratransgenesis. Here, we report on the detection, prevalence and maternal transmission of Asaia among field-caught Anopheles mosquitoes from different eco-geographical localities in Cameroon.
Methods
Study sites and sample collection
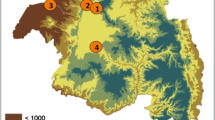
Mosquitoes were collected in eight localities in Cameroon, namely Gounougou (Northern Region 9°03′00″ N, 13°43′59″ E), Tibati (Adamaoua Region, 6°28′00″ N, 12°38′00″ E), Mibellon (Adamaoua Region, 6°46′ N, 11°70′ E), Mangoum (West Region, 5°28′60″ N, 10°34′0″ E), Bankeng (Centre Region, 4°40′26.4″ N, 12°22′30″ E), Elon (Centre Region, 4°15′0″ N, 11°37′0″ E), Elende (Centre Region, 3°41′57.27″ N, 11°33′28.46″ E) and Obout (Centre Region, 3°28′17.0″ N, 11°44′09.4″ E) (Fig. 1), from July 2016 to February 2020. The samples from Mibellon, Gounougou and Bankeng were collected in the rainy season, while those from Elende, Elon, Obout, Mangoum and Tibati were collected in the dry season. Indoor resting female mosquitoes were collected using electric aspirators between 06:00 a.m. and 09:00 a.m. following verbal consent from the chief and each household representative. The collected mosquitoes were then transported to the insectary at the Centre for Research in Infectious Diseases (CRID), Yaoundé, where they were morphologically identified following morphological identification keys for Afrotropical anopheline mosquitoes [29].
Map of the sampling sites. The study sites where the samples were collected are represented by stars. The map was constructed for this publication in QGIS 3.14 (https://www.qgis.org/fr/site/index.html) using country and regional boundaries from GADM (https://gadm.org/download_country_v3.html)
DNA extraction and molecular species identification
Total genomic DNA (gDNA) of each adult mosquito was extracted using the Livak method [30]. Following extraction, the concentration and purity of the extracted gDNA were determined using a NanoDrop™ spectrophotometer (Thermo Scientific, Waltham, MA, USA) before storage at −20 °C. Short interspersed nuclear element (SINE)-based polymerase chain reaction (PCR) [31] and cocktail PCR [32] were performed to identify the different species of the An. gambiae complex and An. funestus group, respectively.
Plasmodium infection rates
The detection of Plasmodium infection from each whole mosquito was performed using the TaqMan assay as described previously [33]. In this assay, two probes were used to check for the presence or absence of Plasmodium infection. The first probe, labeled with FAM, detects P. falciparum, and the second probe tagged with HEX detects P. vivax, P. ovale and/or P. malariae. This indicates that the effect of bacteria on Plasmodium development has been based only on the presence versus absence of Plasmodium infection.
Asaia screening and sequencing
The presence of Asaia in the mosquitoes was detected with a diagnostic PCR using Asaia-specific primers 16SF (5′-TGG CGG ACG GGT GAG TAT C-3′) and Asarev (5′-AGC GTC AGT AAT GAG CCA GGT T-3′) [26] to amplify 676 base pairs (bp) of the 16S rRNA gene. The reaction mix was run for 5 min at 95 °C and cycled 35 times through 30 s at 94 °C, 30 s at 60 °C and 1 min at 72 °C. Finally, further extension was performed for 10 min at 72 °C. PCR products were then resolved and visualized on 1.5% agarose gel containing Midori green dye. If bands of the expected size were visible on the gels, the PCR products were cleaned up using Exonuclease I (Exo I) and Shrimp Alkaline Phosphatase (Exo-SAP protocol) according to the New England Biolabs protocol (NEB, Ipswich, MA, USA) and sent for sequencing. Sterile water was used as a negative control.
Assessing the vertical transmission of Asaia
To assess the vertical transmission of Asaia in Anopheles mosquitoes, we investigated the presence of the bacteria in matching F0 female parent and F1 female An. funestus progeny from Elende. Blood-fed adult mosquitoes collected in Elende were transferred individually to paper cups and allowed to feed for 4–5 days on 10% sterile sugar-soaked cotton wool. Once fully gravid, females were allowed to oviposit individually by forced egg laying as described previously [34]. After hatching, larvae were reared to obtain F1 adults in isofamilies. F0 mosquitoes were then tested with their respective F1 for the presence of Asaia. Detection for the presence of Asaia by PCR in F1 mosquitoes was also performed, and the positive samples were sequenced to assess whether the Asaia strains from F0 to F1 individuals were the same. The Asaia sequences from F1 progeny were aligned and compared with the sequences of their respective F0 parents. This was done to confirm the vertical transmission pattern of Asaia.
Phylogenetic analysis
Overall, 60 individual samples were analyzed using the 16S rRNA gene. The sample distribution included 10 An. funestus samples each from Mibellon, Obout, Elon and Elende, 10 An. gambiae samples from Mibellon, four An. gambiae samples from Bankeng, five An. coluzzii samples from Gounougou, and one An. funestus sample from Tibati. The number of samples utilized was based on the number of infected mosquitoes among the various species and the clarity of the sequences. Maximum likelihood phylogenetic trees were constructed using the Asaia-specific 16S rRNA gene target. The taxonomic relationships of the strains obtained from this study were inferred against GenBank sequence isolates. These existing isolates are presented in Additional file 1: Table S1. Additionally, these sequences were aligned with the ClustalW multiple sequence alignment tool in BioEdit software. Neoasaia chiangmaiensis (FJ887939.1), Gluconobacter oxydans (KU255083.1) and Acetobacter tropicalis (JF930138.1) were used as outgroups.
The evolutionary history was inferred by the maximum likelihood method using MEGA X based on the Jukes–Cantor model. The robustness of the individual branches was estimated by bootstrapping with 1000 replicates. All the sequences were deposited in GenBank under the accession numbers MW450601–MW450660. Genetic parameters including the number of nucleotide sequence variants, nucleotide sequence variant diversity (Hd), Tajima’s and Fu’s indexes and nucleotide diversity were computed using DnaSP 5.10.01. The TCS program was used to construct the haplotype network [35].
Statistical analysis
Data were computed in MS Excel and analyzed using R software version 1.1.463. The Chi-square test was used to compare the prevalence of Asaia between different localities and species. The comparison of the prevalence of P. falciparum in Asaia-positive and Asaia-negative individuals was done using Fisher’s exact test.
Results
Prevalence of natural Asaia species and infection rates in Anopheles mosquitoes
Overall, 895 Anopheles mosquitoes belonging to four species, namely An. funestus, An. gambiae, An. arabiensis and An. coluzzii, were screened to determine the prevalence of Asaia sp. The results showed a general prevalence of Asaia sp. up to 43% (95% CI = 40–50%) (Table 1). All the species of Anopheles studied were found to be infected by Asaia, with infection rates of 43%, 48.4%, 33.7% and 20% for An. gambiae, An. funestus, An. coluzzii and An. arabiensis, respectively (Table 2).
The prevalence of Asaia varied according to mosquitoes species. Indeed, Asaia prevalence was significantly higher in An. funestus than in An. coluzzii (χ2 = 3.39, df = 1, P = 0.00028) but was similar to the infection rate of An. gambiae (χ2 = 1.33, df = 1, P > 0.05) and An. arabiensis (χ2 = 3.64, df = 1, P = 0.142). Likewise, the prevalence of Asaia in An. gambiae did not differ from that in An. coluzzii (33.7%) (χ2 = 3.39, df = 1, P = 0.066) or An. arabiensis (χ2 = 2.15, df = 1, P = 0.414). Focusing on the different localities and Anopheles species, the infection rate of Asaia registered in An. funestus from Mibellon was 51.6%, compared to 2.2% from Tibati. The prevalence of Asaia was 0% in An. gambiae from Mangoum, whereas the prevalence was 100% in An. funestus from Obout, Elon and Elende. In An. gambiae populations, 56.4% and 64.9% were Asaia-positive in Mibellon and Bankeng, respectively. Anopheles funestus mosquitoes from Mibellon were found to be more frequently infected by Asaia than those from Tibati (χ2 = 100.54, df = 1, P < 0.001), while An. funestus mosquitoes from Obout were more frequently infected than An. funestus from Mibellon (χ2 = 47.2, df = 1, P = 0.001). On the other hand, the prevalence of Asaia in An. gambiae from Mibellon was comparable to that of An. gambiae from Bankeng (χ2 = 0.47, df = 1, P = 0.490).
According to mosquito collection seasons, out of the 458 An. funestus mosquitoes tested, 336 were collected in the dry season and 122 in the rainy season (Table 3). The prevalence of Asaia during the dry (51.6%) and rainy (47.3%) seasons was the same for An. funestus (χ2 = 0.67, df = 1, P = 0.413), showing that season does not have an impact. Regarding An. gambiae, 131 mosquitoes were collected during the rainy season, with an Asaia infection rate of 58.8%, while only 48 were collected during the dry season, with no infection observed (Table 3). This suggests that the rainy season has a positive effect on the prevalence of Asaia in An. gambiae (χ2 = 47.14, df = 1, P = 0.0001).
Phylogenetic analysis and genetic diversity of Asaia sp.
The 470-bp sequences obtained were subjected to BLAST search in GenBank (NCBI), and all had homology with Asaia sp. The relationship between Asaia sp. and Anopheles mosquitoes, notably An. funestus, An. gambiae and An. coluzzii, revealed a clustering of the same species of Asaia irrespective of the locality or species (Fig. 2a). Comparison of the sequences obtained to the reference revealed close relatedness of Asaia strains in five out of the 10 samples (50%) of An. funestus mosquitoes from Mibellon, and all the An. funestus (100%) from Elende, Elon and Obout. The strains shared close similarity with Asaia strains A. platycodi, A. prunellae, A. siamensis and A. lannensis isolated from An. gambiae from Senegal [21], Asaia strain AE isolated from Ae. aegypti from Italy [23] and other Asaia strains isolated from An. stephensi from Italy [17].
Genetic diversity of fragment of 16S rRNA gene for the detection of Asaia sp. in An. coluzzii, An. gambiae and An. funestus at Mibellon, Tibati, Gounougou, Obout, Elende, Elon and Bankeng. a Phylogenetic tree of the sequences based on the 16S rRNA gene. Asaia sequences of the Anopheles populations were used to construct the phylogenetic tree based on the maximum likelihood method with 1000 bootstrap replicates. The sequences of Asaia were aligned against close reference sequences. The bootstrap values under 50% were discarded. b Nucleotide sequence variant network. Nucleotide sequence variant networks were constructed using Asaia sequences in the TCS program. H1–H7 represent the different nucleotide sequence variants, and each color represents each mosquito species according to locality. c Alignment of nucleotide sequence variant showing the polymorphic sites
Similarly, five out of 10 (50%) An. funestus mosquitoes and one out of the four (25%) An. gambiae from Bankeng were found to be infected by a strain of Asaia sp. that clusters with A. krungthepensis and Asaia strain AGF0 of An. gambiae from Burkina Faso [18]. Moreover, Asaia sp. found in the 10 An. gambiae mosquitoes from Mibellon, one An. coluzzii (20%, 1/5) from Gounougou and An. funestus from Tibati cluster with A. astilbis.
Genetic diversity parameters of Asaia sp. revealed seven distinct nucleotide sequence variants (Fig. 2b), and low genetic diversity (π = 0.00241) and nucleotide sequence variant diversity (Hd = 0.481) (Table 4). The result also showed six variable polymorphic sites (Fig. 2c) with slight nucleotide differences. Depending on the mosquito species, we recorded nucleotide diversity of 0.00128, 0.00278 and 0.00084 for An. funestus, An. gambiae and An. coluzzii, respectively (Table 4). The major nucleotide sequence variant H1 (n = 42, frequency = 70%) was present in three species of mosquitoes originating from four locations (Mibellon, Elon, Elende, Bankeng and Gounougou). This suggests that H1 could be the original nucleotide sequence variant. H3 (n = 11, frequency = 18.3%) was found in An. gambiae from Mibellon and An. funestus from Tibati. The nucleotide sequence variant H2 (n = 1, frequency = 1.6%) was present in one individual An. gambiae sample from Bankeng, while nucleotide sequence variant H4 (n = 1, frequency = 1.6%) was detected in An. coluzzii from Gounougou. H5 (n = 1, frequency = 1.6%), H6 (n = 1, frequency = 1.6%) and H7 (n = 1, frequency = 1.6%) nucleotide sequence variants were present exclusively in An. funestus from Mibellon (Fig. 2b). The overall Tajima’s D statistic (D = −0.29815) and Fu’s F statistics (F = −1.335) used to determine the proximity to neutrality were negative and non-significant for the Asaia strains circulating in the Anopheles population from the different localities (Table 4).
Asaia sp. is vertically transmitted and displays stability of infection in Anopheles mosquitoes
Although Asaia is mainly maternally transmitted, horizontal transmission may occasionally occur in natural conditions. To confirm the vertical transmission in the infected mosquito species, we analyzed the infection status of 651 An. funestus F1 mosquitoes from Elende belonging to 29 isofamilies. We found that 601 F1 mosquitoes were Asaia-positive, with average vertical transmission frequency of 91.5% (range: 42.5–100%) (Fig. 3). In addition to the high transmission of Asaia in F1 mosquitoes, comparative phylogenetic analysis of the Asaia sequences from F0 to their F1 progeny revealed identical clustering (Fig. 4). Moreover, this prevalence of transmission could include both vertical and horizontal transmission because the internal control of testing progeny of Asaia-negative parents in order to exclude horizontal transmission through laboratory rearing was lacking in this study.
Asaia has no effect on Plasmodium infection in An. funestus from Mibellon
We compared the overall P. falciparum infection rates (based on the presence vs. absence of the Plasmodium infection) in An. funestus mosquitoes collected from Mibellon to determine a possible correlation with the presence of Asaia infection. Of the 122 mosquitoes sampled, only 25% had detectable Asaia-Plasmodium co-infections, as opposed to 51% Asaia mono-infections. Furthermore, only 31 out of the 122 (26%) mosquitoes were Plasmodium-positive. There was no association between the prevalence of Plasmodium in Asaia-negative and Asaia-positive groups (χ2 = 1.9, df = 1, P > 0.05) of An. funestus from Mibellon. Thus, Asaia has no effect on the natural Plasmodium infection status of Anopheles mosquitoes in Mibellon (Fig. 5).
Discussion
This study provides an estimate of the prevalence of Asaia in natural populations of Anopheles mosquitoes across different localities of Cameroon. The mosquito species studied here have an overall Asaia prevalence of 43%. This prevalence is lower than the prevalence (70%) reported in Anopheles mosquitoes in Iran [26]. Likewise, it is also lower than the prevalence documented in the Centre Region of Cameroon, where Asaia frequency of 95% was observed in the midguts of Anopheles mosquitoes [28]. This difference could be explained by the seasonal, geographical and environmental variation in microbiota composition in Anopheles mosquitoes from the various study sites [36,37,38].
We also found that all the Anopheles species were infected by Asaia. This correlates with previous studies in the Centre Region of Cameroon where Asaia infection was recorded in An. gambiae and An. coluzzii mosquitoes [28]. This finding also parallels that of a study in Madagascar that demonstrated Asaia infection in An. funestus mosquitoes [27]. The presence of natural Asaia in several Anopheles species could be harnessed to develop a paratransgenic control method against Anopheles vectors responsible for malaria. The fact that Asaia has also been described in other species of mosquitoes such as An. stephensi [17] and Ae. aegypti [23] suggests that Asaia is not restricted to certain mosquito species. Rather, it can be widely distributed among many vector species. Moreover, Asaia has been reported to be stably associated with different mosquito species owing to its vertical and horizontal (through pre-adult and adult mosquito oral feeding) transmission potential. This also accounts for its dominance in the mosquito microbiome [17, 18, 23]. Furthermore, Asaia can be found in different niches, including mosquito breeding sites and also in flower nectars. These ecological segments constitute a suitable habitat for these bacteria [21].
The results obtained show variability in infection rates between localities in different species of Anopheles. This is explained by the heterogeneous routes in transmission including feeding on flowers, breeding site, and horizontal and vertical dissemination events [17, 21, 22]. The highest infection rates of Asaia were observed at Elon, Elende and Obout, which are all located in the forest region. This highest infection rate could be attributed to the abundance of plants and breeding sites. Indeed, it has been demonstrated that flower nectar is home to viable strains of Asaia [21].
The clustering of the Asaia strains obtained in this study with other known species of Asaia suggests that the strains of Asaia infecting Anopheles mosquitoes in Cameroon are not novel species. Further investigations are needed to identify each species of Asaia circulating in Anopheles populations across Cameroon. Moreover, the close relatedness with the Asaia strain isolated from other mosquito species from other countries suggests that introducing genetically modified Asaia strains in the Anopheles population will override genetic hurdles of mosquito populations isolated by reproductive barriers. These reproductive barriers often occur in endemic malaria settings and account for limitations observed with vector control strategies. In addition, the overall negative values for both neutrality tests (Tajima’s D and Fu’s Fs tests) based on the 16S rRNA gene indicate an excess of the rare mutations in populations, which suggest a recent population expansion as already demonstrated in a previous study focusing on the genetic diversity of Echinococcus granulosus complex using mitochondrial DNA [39]. Also, analysis of the 16S rRNA gene confirmed that all the strains belong to the genus Asaia, but it was not possible to identify the species, and further analyses such as whole-genome sequencing could be used to better characterize Asaia strains of Anopheles mosquitoes in Cameroon.
This study demonstrated that Asaia-infected females transmit the infection with high frequency (91.5%) to their progeny, suggesting a vertical transmission of Asaia. Moreover, the close relatedness of the Asaia strains between F1 and F0 female mosquitoes suggests that Asaia displays stability of infection and reinforces the vertical transmission potential of the bacteria in Anopheles mosquitoes. This feature is crucial because it offers the possibility for introducing engineered Asaia into mosquito populations in the field which will spread over time and replace the wild-type population. The vertical transmission of Asaia has also been shown in other species of Anopheles [13, 17]. Our results suggest that Asaia is a promising candidate for bacterial engineering for the production of anti-plasmodium effectors. In fact, the high vertical transmission frequency of Asaia is not a surprising event, as it has already been demonstrated in Anopheles mosquitoes [17, 18]. Nevertheless, the high frequency of Asaia transmission recorded in this study could be due to a combination of horizontal and vertical dissemination events during co-feeding of F1 mosquitoes in the laboratory. Given that the presence of Asaia has not been tested in Asaia-negative parents, we cannot exclude the possibility that the DNA sequences detected in this study came from some sort of environmental contamination through sugar meals or other sources. Additional experiments would be of great interest to demonstrate actual infection, for example, showing maternal transmission of the bacteria from Asaia-negative samples and intracellular localization of the sequences. Also, different techniques such as fluorescence in situ hybridization (FISH) could be used to detect Asaia in different tissues (head and thorax, midgut and ovaries). Finally, the estimated rate of transfer of Asaia vertically from parents to progeny reported here also includes the rate at which F1 larvae acquired infection from laboratory sources.
Several distinct microbial-based approaches are currently being investigated [9,10,11]. However, it is unclear whether these approaches are compatible. For example, there is increasing evidence that microbe–microbe interactions influence colonization and abundance within the host [40,41,42], and these parameters are likely critical for successful control strategies. This is true for Asaia and Wolbachia, which appear antagonistic to one another [43, 44]. The recent finding of stable Wolbachia infection in Anopheles mosquitoes reinvigorates the use of this microbe for control of malaria [27, 45]. As such, there need to be careful considerations for the type of microbial control approach to be used in a particular region given that it may impede future control strategies.
Asaia has been shown to be a promising candidate for paratransgenic control of malaria. This is as a result of its negative effect on the development of Plasmodium at different stages [27]. Here we focused on the effect of Asaia on Plasmodium in natural populations. The data obtained show that in a field Anopheles population of Mibellon, the presence of Asaia did not affect the infection of mosquitoes by Plasmodium. These conclusions are in accord with the results of a recent study demonstrating that Asaia and Wolbachia do not influence Plasmodium infection in natural populations [27]. However, Bassene et al. showed that a strain of Asaia correlated negatively with Plasmodium infection in wild populations of An. gambiae sensu lato and An. funestus in Senegal [19]. Moreover, experimental infection studies need to be conducted to better evaluate the relationship between Asaia and Plasmodium infection in wild Anopheles mosquitoes. It will also be important to quantify the level of Plasmodium parasites and Asaia sp. in each Anopheles population to better determine this correlation.
Conclusion
Asaia has been presented as a promising candidate for alternative control of malaria. Our study provides preliminary evidence of the circulation of Asaia in malaria vectors in Cameroon. The predominance of a nucleotide sequence variant in all the mosquito species suggests the feasibility of a paratransgenic control approach via the bioengineering of Asaia in malaria vectors. These data provide important initial baseline information towards developing potential strategies by exploring the possibility of utilizing this strategy for malaria control.
Availability of data and materials
The data are part of a wider study of bacteria detection in Anopheles mosquitoes in Cameroon. Data are available from the corresponding author upon reasonable request.
References
WHO. World malaria report 2020: 20 years of global progress and challenges. 2020.
Tonye SGM, Kouambeng C, Wounang R, Vounatsou P. Challenges of DHS and MIS to capture the entire pattern of malaria parasite risk and intervention effects in countries with different ecological zones: the case of Cameroon. Malar J. 2018;17:156.
Diengou NH, Cumber SN, Nkfusai CN, Mbinyui MS, Viyoff VZ, Bede F, et al. Factors associated with the uptake of intermittent preventive treatment of malaria in pregnancy in the Bamenda health districts, Cameroon. Pan Afr Med J Conf. 2020;35:42.
Antonio-Nkondjio C, Ndo C, Njiokou F, Bigoga JD, Awono-Ambene P, Etang J, et al. Review of malaria situation in Cameroon: technical viewpoint on challenges and prospects for disease elimination. Parasites Vectors. 2019;12:501.
Bhatt S, Weiss D, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207.
Fadel AN, Ibrahim SS, Tchouakui M, Terence E, Wondji MJ, Tchoupo M, et al. A combination of metabolic resistance and high frequency of the 1014F kdr mutation is driving pyrethroid resistance in Anopheles coluzzii population from Guinea savanna of Cameroon. Parasites Vectors. 2019;12:263.
Wondji CS, Dabire RK, Tukur Z, Irving H, Djouaka R, Morgan JC. Identification and distribution of a GABA receptor mutation conferring dieldrin resistance in the malaria vector Anopheles funestus in Africa. Insect Biochem Mol Biol. 2011;41:484–91.
Menze BD, Riveron JM, Ibrahim SS, Irving H, Antonio-Nkondjio C, Awono-Ambene PH, et al. Multiple insecticide resistance in the malaria vector Anopheles funestus from Northern Cameroon is mediated by metabolic resistance alongside potential target site insensitivity mutations. PLoS ONE. 2016;11:e0163261.
Coutinho-Abreu IV, Zhu KY, Ramalho-Ortigao M. Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol Int. 2010;59:1–8.
McGraw EA, O’Neill SL. Beyond insecticides: new thinking on an ancient problem. Nat Rev Microbiol. 2013;11:181–93.
Cansado-Utrilla C, Zhao SY, McCall PJ, Coon KL, Hughes GL. The microbiome and mosquito vectorial capacity: rich potential for discovery and translation. Microbiome. 2021;9:111.
Ricci I, Damiani C, Capone A, DeFreece C, Rossi P, Favia G. Mosquito/microbiota interactions: from complex relationships to biotechnological perspectives. Curr Opin Microbiol. 2012;15:278–84.
Capone A, Ricci I, Damiani C, Mosca M, Rossi P, Scuppa P, et al. Interactions between Asaia, Plasmodium and Anopheles: new insights into mosquito symbiosis and implications in malaria symbiotic control. Parasites Vectors. 2013;6:182.
Saldaña MA, Hegde S, Hughes GL. Microbial control of arthropod-borne disease. Mem Inst Oswaldo Cruz. 2017;112:81–93.
Kommanee J, Tanasupawat S, Yukphan P, Thongchul N, Moonmangmee D, Yamada Y. Identification of Acetobacter strains isolated in Thailand based on the phenotypic, chemotaxonomic, and molecular characterizations. Sci Asia. 2012;38:44–53.
Crotti E, Rizzi A, Chouaia B, Ricci I, Favia G, Alma A, et al. Acetic acid bacteria, newly emerging symbionts of insects. Appl Environ Microbiol. 2010;76:6963–70.
Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci. 2007;104:9047–51.
Damiani C, Ricci I, Crotti E, Rossi P, Rizzi A, Scuppa P, et al. Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb Ecol. 2010;60:644–54.
Bassene H, Niang EHA, Fenollar F, Doucoure S, Samb B, Faye O, et al. A pilot study on isolation of Asaia and detecting its co-presence with Plasmodium falciparum in two major malaria vectors in Senegal. J Parasitol Vector Biol. 2019;11:1–9.
Favia G, Ricci I, Marzorati M, Negri I, Alma A, Sacchi L, et al. Bacteria of the genus Asaia: a potential paratransgenic weapon against malaria, in transgenesis and the management of vector-borne disease. Adv Exp Med Biol. 2008;627:49–59.
Bassene H, Fenollar F, Doucoure S, Faye O, Raoult D, Sokhna C, et al. Role of plants in the transmission of Asaia sp., which potentially inhibit the Plasmodium sporogenic cycle in Anopheles mosquitoes. Sci Rep. 2020;10:1–10.
Li F, Hua H, Han Y, Hou M. Plant-mediated horizontal transmission of Asaia between white-backed planthoppers, Sogatella furcifera. Front Microbiol. 2020;11: 593485.
Crotti E, Damiani C, Pajoro M, Gonella E, Rizzi A, Ricci I, et al. Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ Microbiol. 2009;11:3252–64.
Chouaia B, Rossi P, Epis S, Mosca M, Ricci I, Damiani C, et al. Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 2012;12(Suppl. 1):S2.
Mitraka E, Stathopoulos S, Siden-Kiamos I, Christophides GK, Louis C. Asaia accelerates larval development of Anopheles gambiae. Pathog Glob Health. 2013;107:305–11.
Rami A, Raz A, Zakeri S, Dinparast Djadid N. Isolation and identification of Asaia sp. in Anopheles spp. mosquitoes collected from Iranian malaria settings: steps toward applying paratransgenic tools against malaria. Parasites Vectors. 2018;11:367.
Jeffries CL, Lawrence GG, Golovko G, Kristan M, Orsborne J, Spence K, et al. Novel Wolbachia strains in Anopheles malaria vectors from sub-Saharan Africa. Wellcome Open Res. 2018;3:113.
Tchioffo MT, Boissière A, Abate L, Nsango SE, Bayibéki AN, Awono-Ambéné PH, et al. Dynamics of bacterial community composition in the malaria mosquito’s epithelia. Front Microbiol. 2016;6:1500.
Gillies M, Coetzee M. A supplement to the Anophelinae of Africa South of the Sahara. Publ S Afr Inst Med Res. 1987;55:1–143.
Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–34.
Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, Della TA. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163.
Koekemoer L, Kamau L, Hunt R, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66:804–11.
Bass C, Nikou D, Blagborough AM, Vontas J, Sinden RE, Williamson MS, et al. PCR-based detection of Plasmodium in Anopheles mosquitoes: a comparison of a new high-throughput assay with existing methods. Malar J. 2008;7:177.
Morgan JC, Irving H, Okedi LM, Steven A, Wondji CS. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS ONE. 2010;5:e11872.
Crandall MCDPK, Clement M, Posada D. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–60.
Akorli J, Gendrin M, Pels NAP, Yeboah-Manu D, Christophides GK, Wilson MD. Seasonality and locality affect the diversity of Anopheles gambiae and Anopheles coluzzii midgut microbiota from Ghana. PLoS ONE. 2016;11:e0157529.
Muturi EJ, Lagos-Kutz D, Dunlap C, Ramirez JL, Rooney AP, Hartman GL, et al. Mosquito microbiota cluster by host sampling location. Parasites Vectors. 2018;11:468.
Krajacich BJ, Huestis DL, Dao A, Yaro AS, Diallo M, Krishna A, et al. Investigation of the seasonal microbiome of Anopheles coluzzii mosquitoes in Mali. PLoS ONE. 2018;13:e0194899.
Sharma M, Fomda BA, Mazta S, Sehgal R, Singh BB, Malla N. Genetic diversity and population genetic structure analysis of Echinococcus granulosus sensu stricto complex based on mitochondrial DNA signature. PLoS ONE. 2013;8:e82904.
Hegde S, Khanipov K, Albayrak L, Golovko G, Pimenova M, Saldana MA, et al. Microbiome interaction networks and community structure from laboratory-reared and field-collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquito vectors. Front Microbiol. 2018;9:2160.
Kozlova EV, Hegde S, Roundy CM, Golovko G, Saldana MA, Hart CE, et al. Microbial interactions in the mosquito gut determine Serratia colonization and blood-feeding propensity. ISME J. 2021;15:93–108.
Zink SD, Van Slyke GA, Palumbo MJ, Kramer LD, Ciota AT. Exposure to West Nile virus increases bacterial diversity and immune gene expression in Culex pipiens. Viruses. 2015;7:5619–31.
Rossi P, Ricci I, Cappelli A, Damiani C, Ulissi U, Mancini MV, et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasites Vectors. 2015;8:278.
Hughes GL, Dodson BL, Johnson RM, Murdock CC, Tsujimoto H, Suzuki Y, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci. 2014;111:12498–503.
Walker T, Quek S, Jeffries CL, Bandibabone J, Dhokiya V, Bamou R, et al. Stable high-density and maternally inherited Wolbachia infections in Anopheles moucheti and Anopheles demeilloni mosquitoes. Curr Biol. 2021;31:2310–20.
Acknowledgements
We thank the field entomology mosquito collection team and the inhabitants of the respective villages in Cameroon where these data were collected. We thank Mr. Francis Nkemngo and native speakers of English for their assistance in the review and editing of this paper.
Funding
This work was funded by the Medical Research Council, UK, and Global Challenges Research Fund, through the PIIVeC. Grant Number MR/P027873/1.
Author information
Authors and Affiliations
Contributions
MMS and CSW conceived the study. MMS designed the study. CGM, MMS, ANK, MT and BM collected the samples in the field. CGM and MMS performed the experimental analysis. CGM and DNN performed the genetic analysis. CGM and MMS wrote the manuscript with contributions from MOK, GLH, CSW and FN. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Verbal community acceptance was obtained from the village local council representatives. Oral consent for sampling inside houses was obtained from household owners before collection.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
References sequences and accession numbers.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Maffo, C.G.T., Sandeu, M.M., Fadel, A.N. et al. Molecular detection and maternal transmission of a bacterial symbiont Asaia species in field-caught Anopheles mosquitoes from Cameroon. Parasites Vectors 14, 539 (2021). https://doi.org/10.1186/s13071-021-05044-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-05044-2