Abstract
Background
The emergence of tick-borne disease is increasing because of the effects of the temperature rise driven by global warming. In Turkey, 19 pathogens transmitted by ticks to humans and animals have been reported. Based on this, this study aimed to investigate tick-borne pathogens including Hepatozoon spp., Theileria spp., Babesia spp., Anaplasma spp., Borrelia spp., and Bartonella spp. in tick samples (n = 110) collected from different hosts (dogs, cats, cattle, goats, sheep, and turtles) by molecular methods.
Methods
To meet this objective, ticks were identified morphologically at the genus level by microscopy; after DNA isolation, each tick sample was identified at the species level using the molecular method. Involved pathogens were then investigated by PCR method.
Results
Seven different tick species were identified including Rhipicephalus sanguineus, R. turanicus, R. bursa, Hyalomma marginatum, H. anatolicum, H. aegyptium, and Haemaphysalis erinacei. Among the analyzed ticks, Hepatozoon spp., Theileria spp., Babesia spp., and Anaplasma spp. were detected at rates of 6.36%, 16.3%, 1.81%, and 6.36%, respectively while Borrelia spp. and Bartonella spp. were not detected. Hepatozoon spp. was detected in R. sanguineus ticks while Theileria spp., Babesia spp., and Anaplasma spp. were detected in R. turanicus and H. marginatum. According to the results of sequence analyses applied for pathogen positive samples, Hepatozoon canis, Theileria ovis, Babesia caballi, and Anaplasma ovis were identified.
Conclusion
Theileria ovis and Anaplasma ovis were detected for the first time to our knowledge in H. marginatum and R. turanicus collected from Turkey, respectively. Also, B. caballi was detected for the first time to our knowledge in ticks in Turkey.
Graphical Abstract

Similar content being viewed by others
Background
Ticks are obligate blood-sucking arachnid ectoparasites belonging to the Ixodida suborder that feeds on a wide variety of wild and domestic vertebrates excluding fish [1,2,3]. To date, 896 tick species identified are classified into three families: Ixodidae (hard ticks, 702 species), Argasidae (soft ticks, 193 species), and Nuttalliellidae (Nuttalliella namaqua, 1 species) [3]. After mosquitoes, ticks are the second most common pathogen vectors worldwide [1, 4, 5], and tick-borne diseases (TBDs) are increasingly threatening animal and human health along with causing economic losses [3, 6].
To date, 19 tick-borne pathogens have been reported in Turkey [7]. Among them, causative agents of hepatozoonosis, theileriosis, babesiosis, anaplasmosis, bartonellosis, and Lyme disease are frequently investigated in both ticks and their vertebrate hosts in Turkey and other countries. Hepatozoon, an apicomplexan intraerythrocytic genus of parasites represented by > 300 species, and belonging to the Hepatozoidae family, is commonly detected in tetrapod vertebrates and numerous hematophagous invertebrates [8, 9]. Among these species, H. canis and H. felis cause canine and feline hepatozoonosis, respectively, which are important in the veterinary field [10]. In Turkey, H. canis was detected in R. sanguineus collected from dogs and Haemaphysalis parva collected from red foxes [11, 12]. Theileria is another apicomplexan parasite that belongs to the order Piroplasmida, and it infects many domestic and wild ruminant animals. Theileria parva, T. annulata, and T. orientalis infect cattle [10, 13], while T. lestoquardi, T. luwenshuni, T. separata, and T. ovis infect small ruminants such as sheep and goats [14, 15]. In Turkey, T. ovis has been detected in R. bursa, R. turanicus, and R. sanguineus collected from sheep and goats [16, 17]. Babesia is another important apicomplexan parasite that belongs to the order Piroplasmida, which is transmitted by ticks and infects the red blood cells of various mammals such as cattle, sheep, horses, dogs, and rodents [10]. The vectors of Babesia parasites transmitted to humans and animals are the Ixodid tick species [18]. Babesia caballi together with T. equi causes equine piroplasmosis disease in horses and is transmitted by Dermacentor, Rhipicephalus, and Hyalomma [19,20,21]. Among bacterial pathogens, Anaplasma is a gram-negative bacterium that is a member of the family Anaplasmataceae in the order Rickettsiales, and it affects human and animal health by causing tick-borne diseases with species such as A. phagocytophilum, A. centrale, A. marginale, A. bovis, A. platys, and A. ovis [22,23,24]. Although A. ovis infects sheep, goats, and wild ruminants in Africa, Asia, Europe, and the US, and shows less pathogenicity than other Anaplasma species, it is the main species causing anaplasmosis in small ruminants with subclinical infections [25, 26]. Borrelia (Borreliella) is another bacterial pathogen in the Spirochaetaceae family that causes tick-borne Lyme borreliosis disease and is transmitted by Ixodes tick vectors in Europe, the Far East, and North America [27,28,29]. Borrelia burgdorferi (s.l.) includes 20 species, and 9 of them are known as human and animal pathogens [29]. In addition to ticks, Bartonella, which is another bacterial pathogen, can be transmitted by fleas and lice and can infect domestic and wild mammals, including humans. Twenty-three species have been identified, and 13 of them have been found to be associated with human diseases [30, 31]. Among Bartonella species, B. henselae and B. clarridgeiae cause cat-scratch disease, while B. quintana causes trench fever disease [31, 32]. In Turkey, B. henselae and B. clarridgeiae have been reported in domestic cats [33].
In this study, we aimed to investigate tick-borne pathogens including Hepatozoon spp., Theileria spp., Babesia spp., Anaplasma spp., Borrelia spp., and Bartonella spp. in tick samples collected from different hosts from the İzmir, Aydın, Şanlıurfa, and Siirt provinces of Turkey by PCR method.
Methods
Morphological identification of tick samples
Tick samples (n = 110) were collected from a variety of host organisms [dog (n = 46), cat (n = 1), cattle (n = 14), goat (n = 11), sheep (n = 35), turtles (n = 3)] in four provinces [İzmir (n = 57), Aydın (n = 7), Şanlıurfa (n = 16), Siirt = 30] of Turkey between May 2016 and July 2020 (Figs. 1 and 2). Each tick sample was collected from a different host, and all of them were selected randomly. The collections were conducted with the owners’ consent. Tick samples attached to the animals were collected, and most of them were fully engorged but some were semi-engorged. Also, all of ticks were adults except for a single tick sample that was a nymph (sample no. 62). Ticks were removed by a sterile pair of forceps and kept in separate vials filled with 70% ethanol and morphologically identified at the genus level under a stereo-microscope using identification keys as described [34,35,36].
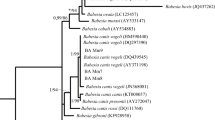
A phylogenetic tree of tick samples based on the 16S rDNA. Each tick sample in the phylogenetic tree clustered with the sequence of the reference tick species downloaded from NCBI. Colored dots represent host organisms from which tick samples were collected. Black, pink, red, orange, blue, and green colored dots represent dog (Canis lupus familiaris), cat (Felis catus), sheep (Ovis aries), goat (Capra hircus), cattle (Bos taurus), and tortoise (Testudo graeca) hosts, respectively. Each colored branch represents a cluster of ticks in the phylogenetic tree. Pathogens are given in black boxes near the cluster of tick species in which they were detected. Silhouette images for the tick and host organisms were retrieved from PhyloPic (http://phylopic.org)
Molecular identification of tick samples
DNA was isolated from each tick sample by a commercial DNA extraction kit (Qiagen) according to the protocol of the manufacturer. Before DNA isolation, each tick sample that was kept in ethanol was rinsed twice with sterile distilled water, dried on a sterile filter, and transferred to a 1.5-ml tube containing tissue lysis buffer and proteinase K. Later, the ticks were cut in to small pieces using a sterile scalpel and homogenized by a sterile micro-pestle. The obtained homogenate was incubated at 55 °C until all small tissue pieces had been lysed, and then the DNA isolation protocol was continued.
In the molecular identification of tick samples, the mitochondrial 16S rDNA was amplified using 5′-CCGGTCTGAACTCAGATCAAGT-3′ and 5′-CTGCTCAATGATTTTTTAAATTGCTGTGG-3 primers as described by Mendell et al. [37]. The 25 µl PCR reaction included 2 µl template DNA, 1 µl primers (0.4 mM each), 12.5 µl DreamTaq PCR Master Mix (2×, Thermo Scientific), and 8.5 µl nuclease-free water. The PCR was performed using the following protocol: 5 min initial denaturation step at 95 °C, followed by 10 cycles of 1 min at 92 °C, 1 min at 48 °C, and 90 s at 72 °C; 32 cycles of 1 min at 92 °C, 35 s at 54 °C, and 90 s at 72 °C; and a final extension of 7 min at 72 °C. All PCR products were separated in 1% agarose gel, stained with nucleic acid dye (SafeView) and visualized.
Molecular investigation of pathogens by PCR
For detection of Hepatozoon spp., Babesia/Theileria spp., Anaplasma spp., Borrelia spp., and Bartonella spp., the used PCR methods, target genes, primers, expected PCR product sizes, and reaction and amplification conditions are shown in Table 1. For each PCR, DreamTaq PCR Master Mix (2×, Thermo Scientific) was used.
Sequencing
PCR products belonging to ticks and positive pathogen samples were sequenced, and the generated sequences were aligned and edited by MEGA7.0 software. Later, a BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed by comparing with the reference samples in National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) to identify ticks and tick-borne pathogen species.
Phylogenetic analysis
For species identification of tick samples and pathogens, phylogenetic analysis was conducted after a model test selection. Accordingly, the mitochondrial 16S rDNA gene region (375 bp) of tick samples was aligned in MEGA7.0, and the phylogenetic tree was reconstructed by the maximum Likelihood method using the Kimura 2 (K2) model with 1000 bootstrap replications. As reference 16S rDNA sequences, R. sanguineus (KR870984), R. turanicus (KY583080), R. bursa (MK620873), H. marginatum (MH663983), H. anatolicum (MH447260), H. aegyptium (KR870970), and Hae. erinacei (KR870975) deposited in NCBI database (https://www.ncbi.nlm.nih.gov) were used. The 18S rDNA gene region (613 bp) of H. canis samples was aligned in MEGA7.0, and the phylogenetic tree was reconstructed by the minimum evolution method using the Tamura 3-parameter (T92) model with 1000 bootstrap replications. As reference 18S rDNA sequences, H. canis (MH922768.1), H. felis (AB771571.1), and Toxoplasma gondii (L24381.1; as an outgroup) deposited in NCBI database (https://www.ncbi.nlm.nih.gov) were used. 18S rDNA gene region (1034 bp) belonging to T. ovis and B. caballi samples was aligned in MEGA7.0, and the phylogenetic tree was reconstructed by the maximum likelihood method using the Tamura three-parameter gamma distribution (T92+G) model with 1000 bootstrap replications. As reference 18S rDNA sequences, T. ovis (FJ603460.1), T. annulata (KF429800.1), T. parva (L02366.1), B. caballi (EU888901.1), B. bigemina (KM046917.1), B. microti (LC127372.1), and Plasmodium falciparum (JQ627152.1; as an outgroup) species deposited in the NCBI database (https://www.ncbi.nlm.nih.gov) were used. Msp4 gene sequence (321 bp) of A. ovis sample was aligned with reference sequences, and a phylogenetic tree was reconstructed by the maximum likelihood method using the Kimura-2 (K2) model with 1000 bootstrap replications. As reference Msp4 sequences, A. ovis (KU497712.1), A. phagocytophilum (KC847317.1), and A. marginale (KU497715.1) species deposited in NCBI database (https://www.ncbi.nlm.nih.gov) were used.
Statistical analysis
Molecular detection proportions between the Aegean region and Southeastern Anatolia region as well as between tick species were computed, and comparison of the proportions was performed by the chi-square test using the PASW Statistics 18 software. Statistically significant differences were determined at P < 0.05.
Results
Morphological and molecular identification of ticks
Among 110 tick samples, morphological analysis showed that there were three genera including Rhipicephalus (n = 71; 64.5%), Hyalomma (n = 37; n = 33.6%), and Haemaphysalis (n = 2; 1.8%). According to NCBI-BLAST and phylogenetic analysis results, R. sanguineus (n = 27; 24.5%), R. turanicus (n = 31; 28.1%), R. bursa (n = 13; 11.8%), H. marginatum (n = 30; 27.2%), H. anatolicum (n = 4; 3.6%), H. aegyptium (n = 3; 2.72%), and Hae. erinacei (n = 2; 1.8%) were identified (Fig. 1).
Molecular detection rates of involved pathogens
At least one pathogen DNA was amplified from 31 of 110 (28.1%) tick samples studied. Three tick samples (3/110; 2.72%) including R. turanicus and H. marginatum species were detected to carry DNA of at least two different pathogens. Tick species that were detected to harbor the pathogen DNA were R. turanicus, H. marginatum, and R. sanguineus. The detection rates of pathogen DNA were 25.9% (7/27), 32.2% (10/31), and 46.6% (14/30) for R. sanguineus, R. turanicus, and H. marginatum, respectively. Hepatozoon spp. was detected in R. sanguineus collected from dogs in İzmir, and the detection rate of Hepatozoon spp. DNA was 6.36% (7/110). Six of them were successfully sequenced and identified as H. canis (Table 2). Theileria spp. was detected in 18 tick samples whereas Babesia spp. in two tick samples. DNA detection rates were 16.3% (18/110) and 1.81% (2/110) for Theileria spp. and Babesia spp., respectively. Both were detected in R. turanicus and H. marginatum collected from sheep (Table 2). Babesia spp. was found in tick samples collected from İzmir whereas Theileria spp. in İzmir as well as Siirt. Among these positive samples, 15 samples were successfully sequenced, and 13 of them were identified as T. ovis, while 2 were identified as B. caballi. Anaplasma spp. were detected in seven tick samples including H. marginatum and R. turanicus collected from sheep in İzmir and Siirt. The detection rate of Anaplasma spp. DNA was 6.36% (7/110) in tick samples analyzed. Among positive samples, a single positive sample detected in R. turanicus was successfully sequenced and identified as A. ovis (Table 2). Borrelia spp. and Bartonella spp. were not detected among tick samples analyzed.
The percent identity rate among H. canis isolates varied from 99.05 to 100%, whereas for T. ovis isolates, it varied from 99.9 to 100%. The percent identity rate was 100% for B. caballi isolates.
The pathogen DNA positivity proportion detected in the Aegean region was statistically significantly higher than in the Southeastern Anatolia region (P < 0.05). Also, no statistically significant relationship was found in terms of the pathogen detection proportion among pathogen-positive tick species (P > 0.05).
Phylogenetic trees
All analyzed species clustered with their own reference sample, and unexpected branches containing different tick species were not observed in the phylogenetic tree (Fig. 1). In addition, although eight tick sequences (samples 63, 64, 65, 66, 67, 68, 69, and 81) clustered in the same proximity to both R. turanicus and R. sanguineus reference species in the phylogenetic tree (Fig. 1), these sequences matched with R. turanicus in the BLAST analysis. Similarly, unexpected or mixed branches containing different species were not observed in phylogenetic trees constructed for each pathogen (Fig. 3).
Discussion
The number of ticks has increased in suitable ecological habitats because of the rising temperature driven by global warming. To date, 19 tick-borne pathogens were reported in Turkey [7]. Although prevalence studies conducted for tick-borne pathogens are present in Turkey (Table 3), the number of these studies is insufficient in the Aegean region of Turkey, and thus the prevalence of tick-borne pathogens is incompletely known in this region. The present study aimed to investigate the important tick-borne pathogens in tick samples collected from four different provinces of Turkey especially involving the Aegean region. For this aim, identification of tick samples was conducted and R. sanguineus (24.5%), R. turanicus (28.1%), and H. marginatum (27.2%) were confirmed as the most prevalent tick species. Previous studies conducted in Turkey demonstrated that R. turanicus and H. marginatum were prevalent, in line with our study except for R. sanguineus [38, 39].
In addition, such important protozoan and bacterial pathogens also have been investigated in this study; 35.9% of tick samples collected from the Aegean region were pathogen DNA-positive whereas 23.9% of ticks collected from the Southeastern Anatolia region were pathogen DNA-positive. Hepatozoon spp. was detected by conventional PCR with a rate of 6.36% in R. sanguineus collected from dogs in İzmir province of Turkey and identified as H. canis (Table 2). Although R. sanguineus is known to be the main vector for H. canis, the vectoring possibilities of other tick species including R. turanicus, Hae. longicornis, Hae. flava, and Amblyomma ovale have been reported [40,41,42]. In Turkey, H. canis was detected by conventional PCR with a prevalence of 33.3% in Hae. parva collected from red foxes in Ankara while it was detected by conventional PCR with a prevalence of 20.58% in R. sanguineus collected from dogs in Diyarbakır [11, 12] (Table 3). In addition to Turkey, H. canis was detected by conventional PCR in adult R. sanguineus species and nymph Ixodes ricinus collected from dogs in Italy with prevalences of 33% and 5%, respectively [43]. These findings show that Hae. parva also can harbor the H. canis in Turkey in addition to the main vector, R. sanguineus.
In this study, Theileria spp. was detected by nested PCR with a rate of 16.3% in R. turanicus and H. marginatum collected from sheep in İzmir and Siirt provinces and identified as T. ovis (Table 2). Also, the present study is the first reporting the detection of T. ovis in H. marginatum in addition to R. bursa, R. turanicus, and R. sanguineus in Turkey. For example, T. ovis was detected by conventional PCR with a prevalence of 19.27% in R. bursa collected from sheep and goats in Elazig province of Turkey [16] (Table 3), and it was known that T. ovis was the most common Theileria species in small ruminants and its vector was R. bursa in the Mediterranean basin and Turkey [14, 16, 44, 45]. However, another work conducted in the Black Sea region of Turkey (Bolu, Kastamonu, Çorum, Samsun, Tokat, Giresun, and Bayburt) detected T. ovis by conventional PCR in R. turanicus and R. sanguineus in addition to R. bursa collected from sheep and goats with prevalences of 2.27%, 1.47%, and 2.37%, respectively [17] (Table 3).
In this study, Babesia spp. was detected by nested PCR with a rate of 1.81% in H. marginatum and R. turanicus collected from sheep in İzmir and identified as B. caballi. This was the second important first report in this study, demonstrating the presence of B. caballi in two different tick species in Turkey (Table 2). A previous study conducted in Erzurum province of Turkey screened B. caballi by multiplex PCR in horses but was not detected [20] while another study detected it with a prevalence of 3% by PCR in horses in Ankara [46]. Also, no report has detected B. caballi in ticks in Turkey. Apart from Turkey, B. caballi was detected by conventional PCR with a prevalence of 7.6% in R. evertsi evertsi from horses and donkeys in Nigeria while it was detected by nested PCR with a prevalence of 12.9% in Dermacentor nuttalli from horses in Mongolia [47, 48]. Also, B. caballi was detected with a prevalence of 14.55% in 2017 while it was detected with a prevalence of 27.59% in 2018 by real-time PCR in R. bursa collected from vegetation in Italy [21].
In this study, Anaplasma spp. was detected by nested PCR with a rate of 6.36% in H. marginatum and R. turanicus collected from sheep in the Siirt and İzmir provinces of Turkey, and only one sample detected in R. turanicus could be sequenced and identified as A. ovis (Table 2). This was the third important first report in this study demonstrating the presence of A. ovis in R. turanicus in Turkey (Table 2). In Turkey, A. ovis was not detected by conventional PCR in ticks collected from several animals in Ankara [49] (Table 3). Similarly, a previous work comprising Ankara, Bolu, and Kırşehir provinces did not detect A. ovis by conventional PCR in several ticks collected from animals including cattle, sheep, goats, and dogs [50] (Table 3). However, another study detected A. ovis by PCR with a prevalence of 0.82 (2/242) in R. sanguineus collected from sheep in Ankara [51] (Table 3). Apart from Turkey, A. ovis was detected by conventional PCR with a prevalence of 2.3% in Hae. longicornis collected from sheep in China while it was detected by conventional PCR with a prevalence of 2.4% in D. nuttalli collected from sheep and goats in Mongolia [23, 52]. Also, A. ovis was detected by real-time PCR with a prevalence of 20.3% in R. bursa collected from goats in France [26].
Interestingly, Borrelia spp. and Bartonella spp. were not detected among tick samples analyzed in the present study. Indeed, Borrelia spp. has been detected at high prevalence rates in tick samples analyzed in previous studies conducted in Turkey. For example, Borrelia spp. was detected by nested PCR with a prevalence of 20% in R. turanicus collected from boars in Ankara [49]. A different study conducted in İstanbul detected Borrelia spp. by conventional PCR with prevalence rates of 44% and 39% in adult and nymph forms of H. aegyptium, respectively, which were collected from tortoises (Testudo graeca) [53]. Also, B. burgdorferi was detected by nested PCR with prevalence rates of 38.7% and 11.4% in İstanbul and Kırklareli provinces, respectively [54]. In addition, to date, the molecular prevalence of B. henselae has only been reported in domestic cats in Turkey [7, 33], and no molecular prevalence study is available on the existence of Bartonella spp. in ticks.
This study has some limitations. First is the small sample size. A more comprehensive study with more samples can enable reaching a more precise pathogen detection rate as well as detect the Borrelia spp. and Bartonella spp. in these study areas. The other is the use of a single molecular diagnostic method. In addition to this method, the use of microscopy and culture methods or additional molecular methods such as RNA analyses indicating pathogen-related gene expression can indicate that ticks were infected with the detected pathogens and these pathogens could be transmitted by ticks to vertebrate hosts.
Conclusions
Hepatozoon canis, T. ovis, B. caballi, and A. ovis were detected among tick samples studied contrary to Borrelia spp. and Bartonella spp. Overall, at least one pathogen DNA was detected in 28.1% of tick samples studied. Among these findings, three of them are of great importance for studies conducted in this field in Turkey. The first one is the first detection of T. ovis in H. marginatum whereas the second one is the first detection of A. ovis in Rh. turanicus, and the third one is the first detection of B. caballi in both H. marginatum and R. turanicus species, which have veterinary and medical importance.
Availability of data and materials
All sequences obtained from pathogens were deposited into GenBank (National Center for Biotechnology Information Search database). Provided GenBank Accession numbers are as follows: MW810676, MW810677, MW810626, MW810627, MW810628, MW810629, MW810630, MW810631, MW810474, MW810475, MW810476, MW810477, MW810478, MW810479, MW810480, MW810481, MW810482, MW810483, MW810484, MW810485, MW810486, and MW821793.
References
Nava S, Guglielmone AA, Mangold AJ. An overview of systematics and evolution of ticks. Front Biosci. 2009;14:2857–77. https://doi.org/10.2741/3418.
Kaufman WR. Ticks: physiological aspects with implications for pathogen transmission. Ticks Tick Borne Dis. 2010;1:11–22. https://doi.org/10.1016/j.ttbdis.2009.12.001.
Wang T, Zhang S, Pei T, Yu Z, Liu J. Tick mitochondrial genomes: structural characteristics and phylogenetic implications. Parasit Vectors. 2019;12:451. https://doi.org/10.1186/s13071-019-3705-3.
Fivaz B, Petney T, Horak I. Tick vector biology: medical and veterinary aspects. Berlin: Springer Science & Business Media; 1992.
de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. 2008;13:6938–46. https://doi.org/10.2741/3200.
Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. 2004;129:3–14. https://doi.org/10.1017/s0031182004005967.
Inci A, Yildirim A, Duzlu O, Doganay M, Aksoy S. Tick-borne diseases in Turkey: a review based on one health perspective. PLoS Negl Trop Dis. 2016;10:e0005021. https://doi.org/10.1371/journal.pntd.0005021.
Smith TG. The genus Hepatozoon (Apicomplexa: Adeleina). J Parasitol. 1996;82:565–85.
Baneth G, Samish M, Shkap V. Life cycle of Hepatozoon canis (Apicomplexa: Adeleorina: Hepatozoidae) in the tick Rhipicephalus sanguineus and domestic dog (Canis familiaris). J Parasitol. 2007;93:283–99. https://doi.org/10.1645/GE-494R.1.
Masatani T, Hayashi K, Andoh M, Tateno M, Endo Y, Asada M, et al. Detection and molecular characterization of Babesia, Theileria, and Hepatozoon species in hard ticks collected from Kagoshima, the southern region in Japan. Ticks Tick Borne Dis. 2017;8:581–7. https://doi.org/10.1016/j.ttbdis.2017.03.007.
Aktas M, Özübek S, Ipek DNS. Molecular investigations of Hepatozoon species in dogs and developmental stages of Rhipicephalus sanguineus. Parasitol Res. 2013;112:2381–5. https://doi.org/10.1007/s00436-013-3403-6.
Orkun Ö, Nalbantoğlu S. Hepatozoon canis in Turkish red foxes and their ticks. Vet Parasitol Reg Stud Rep. 2018;13:35–7. https://doi.org/10.1016/j.vprsr.2018.03.007.
Morrison WI, McKeever DJ. Current status of vaccine development against Theileria parasites. Parasitology. 2006;133:169–87. https://doi.org/10.1017/S0031182006001867.
Altay K, Aktaş M, Dumanli N. Theileria infections in small ruminants in the east and southeast Anatolia. Turkiye Parasitol Derg. 2007;31:268–71.
Azmi K, Al-Jawabreh A, Abdeen Z. Molecular detection of Theileria ovis and Theleiria equi in livestock from Palestine. Sci Rep. 2019;9:11557. https://doi.org/10.1038/s41598-019-47965-0.
Aktas M, Altay K, Dumanli N. PCR-based detection of Theileria ovis in Rhipicephalus bursa adult ticks. Vet Parasitol. 2006;140:259–63. https://doi.org/10.1016/j.vetpar.2006.04.005.
Aydin MF, Aktas M, Dumanli N. Molecular identification of Theileria and Babesia in ticks collected from sheep and goats in the Black Sea region of Turkey. Parasitol Res. 2015;114:65–9. https://doi.org/10.1007/s00436-014-4160-x.
Teal AE, Habura A, Ennis J, Keithly JS, Madison-Antenucci S. A new real-time PCR assay for improved detection of the parasite Babesia microti. J Clin Microbiol. 2012;50:903–8. https://doi.org/10.1128/JCM.05848-11.
Englund L, Pringle J. New diseases and increased risk of diseases in companion animals and horses due to transport. Acta Vet Scand Suppl. 2003;100:19–25.
Guven E, Avcioglu H, Deniz A, Balkaya İ, Abay U, Yavuz Ş, et al. Prevalence and molecular characterization of Theileria equi and Babesia caballi in jereed horses in Erzurum, Turkey. Acta Parasitol. 2017;62:207–13. https://doi.org/10.1515/ap-2017-0025.
Romiti F, Magliano A, Antognetti V, Manna G, Cersini A, Scicluna MT, et al. Investigation of Ixodid ticks as vectors of Babesia caballi and Theileria equi (Protozoa: Apicomplexa) in central Italy. J Vector Ecol. 2020;45:25–31. https://doi.org/10.1111/jvec.12370.
Aktas M, Vatansever Z, Altay K, Aydin MF, Dumanli N. Molecular evidence for Anaplasma phagocytophilum in Ixodes ricinus from Turkey. Trans R Soc Trop Med Hyg. 2010;104:10–5. https://doi.org/10.1016/j.trstmh.2009.07.025.
Peng Y, Qi M, Jian F, Wang J, Lv Y, Wei J, et al. Molecular identification of tick-borne pathogens in tick Haemaphysalis longicornis from sheep in Henan, China. Turk J Vet Anim Sci. 2017;41:51–5. https://doi.org/10.3906/vet-1507-103.
Wang J, Zhang Y, Wang X, Cui Y, Yan Y, Wang R, et al. A loop-mediated isothermal amplification assay targeting 16S rRNA gene for rapid detection of Anaplasma phagocytophilum infection in sheep and goats. J Parasitol. 2017;103:187–92. https://doi.org/10.1645/16-158.
Liu Z, Peasley AM, Yang J, Li Y, Guan G, Luo J, et al. The Anaplasma ovis genome reveals a high proportion of pseudogenes. BMC Genomics. 2019;20:69. https://doi.org/10.1186/s12864-018-5374-6.
Cabezas-Cruz A, Gallois M, Fontugne M, Allain E, Denoual M, Moutailler S, et al. Epidemiology and genetic diversity of Anaplasma ovis in goats in Corsica, France. Parasit Vectors. 2019;12:3. https://doi.org/10.1186/s13071-018-3269-7.
Kar S, Yilmazer N, Midilli K, Ergin S, Alp H, Gargili A. Presence of the zoonotic Borrelia burgdorferi sl. and Rickettsia spp. in the ticks from wild tortoises and hedgehogs. Clin Exp Health Sci. 2011;1:166–70.
Yang J, Guan G, Niu Q, Liu Z, Li Y, Liu J, et al. Development and application of a loop-mediated isothermal amplification assay for rapid detection of Borrelia burgdorferi s. l. in ticks. Transbound Emerg Dis. 2013;60:238–44. https://doi.org/10.1111/j.1865-1682.2012.01335.x.
Mendoza-Roldan JA, Colella V, Lia RP, Nguyen VL, Barros-Battesti DM, Iatta R, et al. Borrelia burgdorferi (sensu lato) in ectoparasites and reptiles in southern Italy. Parasit Vectors. 2019;12:35. https://doi.org/10.1186/s13071-019-3286-1.
Diaz MH, Bai Y, Malania L, Winchell JM, Kosoy MY. Development of a novel genus-specific real-time PCR assay for detection and differentiation of Bartonella species and genotypes. J Clin Microbiol. 2012;50:1645–9. https://doi.org/10.1128/JCM.06621-11.
Staggemeier R, Pilger DA, Spilki FR, Cantarelli VV. Multiplex SYBR® green-real time PCR (qPCR) assay for the detection and differentiation of Bartonella henselae and Bartonella clarridgeiae in cats. Rev Inst Med Trop Sao Paulo. 2014;56:93–5. https://doi.org/10.1590/S0036-46652014000200001.
Morozova OV, Cabello FC, Dobrotvorsky AK. Semi-nested PCR detection of Bartonella henselae in Ixodes persulcatus ticks from Western Siberia, Russia. Vector Borne Zoonotic Dis. 2004;4:306–9. https://doi.org/10.1089/vbz.2004.4.306.
Celebi B, Kilic S, Aydin N, Tarhan G, Carhan A, Babur C. Investigation of Bartonella henselae in cats in Ankara, Turkey. Zoonoses Public Health. 2009;56:169–75. https://doi.org/10.1111/j.1863-2378.2008.01170.x.
Walker JB, Keirans JE, Horak IG. The Genus Rhipicephalus (Acari, Ixodidae): a guide to the brown ticks of the world. revised. Cambridge: Cambridge University Press; 2000.
Estrada-Peña A, Bouattour A, Camicas JL, Walker AR. Ticks of domestic animals in the Mediterranean region. Zaragoza: University of Zaragoza; 2004.
Apanaskevich DA, Horak IG. The genus Hyalomma Koch, 1844: V. Re-evaluation of the taxonomic rank of taxa comprising the H.(Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int J Acarol. 2008;34:13–42. https://doi.org/10.1080/01647950808683704.
Mendell NL, Reynolds ES, Blanton LS, Hermance ME, Londoño AF, Hart CE, et al. Detection of Rickettsiae, Borreliae, and Ehrlichiae in ticks collected from Walker County, Texas, 2017–2018. Insects. 2019;10:315. https://doi.org/10.3390/insects10100315.
Karasartova D, Gureser AS, Gokce T, Celebi B, Yapar D, Keskin A, et al. Bacterial and protozoal pathogens found in ticks collected from humans in Corum province of Turkey. PLoS Negl Trop Dis. 2018;12:e0006395. https://doi.org/10.1371/journal.pntd.0006395.
Keskin A, Keskin A, Bursali A, Tekin S. Ticks (Acari: Ixodida) parasitizing humans in Corum and Yozgat provinces, Turkey. Exp Appl Acarol. 2015;67:607–16. https://doi.org/10.1007/s10493-015-9966-4.
Otranto D, Dantas-Torres F, Weigl S, Latrofa MS, Stanneck D, Decaprariis D, et al. Diagnosis of Hepatozoon canis in young dogs by cytology and PCR. Parasit Vectors. 2011;4:55. https://doi.org/10.1186/1756-3305-4-55.
Giannelli A, Lia RP, Annoscia G, Buonavoglia C, Lorusso E, Dantas-Torres F, et al. Rhipicephalus turanicus, a new vector of Hepatozoon canis. Parasitology. 2017;144:730–7. https://doi.org/10.1017/S003118201600250X.
de Castro DL, Rubini AS, dos Santos PK, Metzger B, de Paula Antunes JM, Martins TF, et al. Investigation of tick vectors of Hepatozoon canis in Brazil. Ticks Tick Borne Dis. 2013;4:542–6. https://doi.org/10.1016/j.ttbdis.2013.07.006.
Giannelli A, Ramos RAN, Dantas-Torres F, Mencke N, Baneth G, Otranto D. Experimental evidence against transmission of Hepatozoon canis by Ixodes ricinus. Ticks Tick Borne Dis. 2013;4:391–4. https://doi.org/10.1016/j.ttbdis.2013.03.001.
Sevinc F, Xuan X. Major tick-borne parasitic diseases of animals: a frame of references in Turkey. Eurasian J Vet Sci. 2015;31:132–42.
Uilenberg G. International collaborative research: significance of tick-borne hemoparasitic diseases to world animal health. Vet Parasitol. 1995;57:19–41. https://doi.org/10.1016/0304-4017(94)03107-8.
Güçlü HZ, Karaer KZ. Detection of Babesia caballi (Nuttall, 1910) and Theileria equi (Syn. Babesia equi, Laveran, 1901) by the polymerase chain reaction (PCR) in show and sport horses in the region of Ankara. Türkiye Parazitol Derg. 2007;31:89–93.
Onyiche TE, Taioe MO, Ogo NI, Sivakumar T, Biu AA, Mbaya AW, et al. Molecular evidence of Babesia caballi and Theileria equi in equines and ticks in Nigeria: prevalence and risk factors analysis. Parasitology. 2020;147:1238–48. https://doi.org/10.1017/S0031182020000992.
Battsetseg B, Xuan X, Ikadai H, Bautista JLR, Byambaa B, Boldbaatar D, et al. Detection of Babesia caballi and Babesia equi in Dermacentor nuttalli adult ticks. Int J Parasitol. 2001;31:384–6. https://doi.org/10.1016/s0020-7519(01)00120-5.
Orkun Ö, Çakmak A. Molecular identification of tick-borne bacteria in wild animals and their ticks in Central Anatolia, Turkey. Comp Immunol Microbiol Infect Dis. 2019;63:58–65. https://doi.org/10.1016/j.cimid.2018.12.007.
Orkun Ö. Molecular investigation of the natural transovarial transmission of tick-borne pathogens in Turkey. Vet Parasitol. 2019;273:97–104. https://doi.org/10.1016/j.vetpar.2019.08.013.
Aktas M, Altay K, Dumanli N, Kalkan A. Molecular detection and identification of Ehrlichia and Anaplasma species in ixodid ticks. Parasitol Res. 2009;104:1243–8. https://doi.org/10.1007/s00436-009-1377-1.
Enkhtaivan B, Narantsatsral S, Davaasuren B, Otgonsuren D, Amgalanbaatar T, Uuganbayar E, et al. Molecular detection of Anaplasma ovis in small ruminants and ixodid ticks from Mongolia. Parasitol Int. 2019;69:47–53. https://doi.org/10.1016/j.parint.2018.11.004.
Güner ES, Hashimoto N, Kadosaka T, Imai Y, Masuzawa T. A novel, fast-growing Borrelia sp. isolated from the hard tick Hyalomma aegyptium in Turkey. Microbiology (Reading). 2003;149:2539–44. https://doi.org/10.1099/mic.0.26464-0.
Sen E, Uchishima Y, Okamoto Y, Fukui T, Kadosaka T, Ohashi N, et al. Molecular detection of Anaplasma phagocytophilum and Borrelia burgdorferi in Ixodes ricinus ticks from Istanbul metropolitan area and rural Trakya (Thrace) region of north-western Turkey. Ticks Tick Borne Dis. 2011;2:94–8. https://doi.org/10.1016/j.ttbdis.2011.03.004.
Cardoso L, Cortes HC, Eyal O, Reis A, Lopes AP, Vila-Viçosa MJ, et al. Molecular and histopathological detection of Hepatozoon canis in red foxes (Vulpes vulpes) from Portugal. Parasit Vectors. 2014;7:113. https://doi.org/10.1186/1756-3305-7-113.
Yang J, Liu Z, Niu Q, Liu J, Han R, Liu G, et al. Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasit Vectors. 2016;9:603. https://doi.org/10.1186/s13071-016-1886-6.
Wills MKB, Kirby AM, Lloyd VK. Detecting the lyme disease spirochete, Borrelia burgdorferi, in ticks using nested PCR. J Vis Exp. 2018;132:56471. https://doi.org/10.3791/56471.
Hercik K, Hasova V, Janecek J, Branny P. Molecular evidence of Bartonella DNA in ixodid ticks in Czechia. Folia Microbiol (Praha). 2007;52:503–9. https://doi.org/10.1007/BF02932111.
Acknowledgements
Not applicable.
Funding
This study was supported by a project given by the Ege University Scientific Research Projects Coordination Unit (Project number: TGA-2020-21040) to H.C.
Author information
Authors and Affiliations
Contributions
Conceptualization: HC, and CÜ; Methodology: HC, and CÜ; Formal analysis and investigation: AEK, SEA, MG, HC, ÇK, TT, SD, and MK; Writing-original draft preparation: HC; Writing-review and editing: HC, CÜ, MD, TT, SD, ADD, GA, and AYG; Funding acquisition: HC; Supervision: HC, and CÜ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Köseoğlu, A.E., Can, H., Güvendi, M. et al. Molecular investigation of bacterial and protozoal pathogens in ticks collected from different hosts in Turkey. Parasites Vectors 14, 270 (2021). https://doi.org/10.1186/s13071-021-04779-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-04779-2