Abstract
Background
Belonging to the anopluran family Echinophthiriidae, Echinophthirius horridus, the seal louse, has been reported to parasitise a broad range of representatives of phocid seals. So far, only a few studies have focused on the vector function of echinophthiriid lice, and knowledge about their role in pathogen transmission is still scarce. The current study aims to investigate the possible vector role of E. horridus parasitising seals in the Dutch Wadden Sea.
Methods
E. horridus seal lice were collected from 54 harbour seals (Phoca vitulina) and one grey seal (Halichoerus grypus) during their rehabilitation period at the Sealcentre Pieterburen, The Netherlands. DNA was extracted from pooled seal lice of individual seals for molecular detection of the seal heartworm Acanthocheilonema spirocauda, the rickettsial intracellular bacterium Anaplasma phagocytophilum, and the cell wall-less bacteria Mycoplasma spp. using PCR assays.
Results
Seal lice from 35% of the harbour seals (19/54) and from the grey seal proved positive for A. spirocauda. The seal heartworm was molecularly characterised and phylogenetically analysed (rDNA, cox1). A nested PCR was developed for the cox1 gene to detect A. spirocauda stages in seal lice. A. phagocytophilum and a Mycoplasma species previously identified from a patient with disseminated ‘seal finger’ mycoplasmosis were detected for the first time, to our knowledge, in seal lice.
Conclusions
Our findings support the potential vector role of seal lice in the transmission of A. spirocauda and reveal new insights into the spectrum of pathogens occurring in seal lice. Studies on vector competence of E. horridus, especially for bacterial pathogens, are essentially needed in the future as these pathogens might have detrimental effects on the health of seal populations. Furthermore, studies on the vector role of different echinophthiriid species infecting a wide range of pinniped hosts should be conducted to extend the knowledge of vector-borne pathogens.
Graphical Abstract

Similar content being viewed by others
Background
Representatives of the family Echinophthiriidae belong to the phthirapteran suborder Anoplura, the sucking lice, and exhibit a host range solely infesting semiaquatic mammals, such as pinnipeds (Otariidae, Phocidae, Odobenidae) and the North American river otter (Lontra canadensis) [1]. Within the Echinophthiriidae, five genera (Antarctophthirus, Lepidophthirus, Echinophthirius, Proechinophthirus, and Latagophthirus) and 13 species are described [1, 2]. While many echinophthiriid species display a strict major host specificity, the seal louse Echinophthirius horridus exhibits the broadest host range among echinophthiriid lice parasitising eight different species of Phocidae, the earless or true seals, with a geographical distribution confined to the Northern Hemisphere, including harbour seals (Phoca vitulina) and grey seals (Halichoerus grypus) [1].
Due to their obligate and permanent hematophagous feeding habits [1], members of the Echinophthiriidae have the potential to play an important role in the epidemiology of vector-borne diseases in free-ranging pinniped populations. Nonetheless, studies on the possible vector function of echinophthiriid species are scarce, but some of them already highlighted their role in potential pathogen transmission (Table 1). The bacterium Salmonella enteritidis was isolated from blood and tissue of Northern fur seals (Callorhinus ursinus) and from the echinophthiriid lice Antarctophthirus callorhini and/or Proechinophthirus fluctus collected from the same individuals. Additionally, these infections were also associated with a mortality event of C. ursinus pups [3]. A Rickettsia species closely related to the human pathogen Rickettsia rickettsi was isolated from P. fluctus collected from Northern fur seals [4]. Bartonella henselae was detected in E. horridus and spleen samples of dissected harbour seals [5]. Southern elephant seal virus (SESV), an Alphavirus, was isolated from Lepidophthirus macrorhini parasitising the Southern elephant seal (Mirounga leonina) [6].
Furthermore, E. horridus has been suggested to serve as the obligate intermediate host for the seal heartworm Acanthocheilonema spirocauda, i.e. A. spirocauda-microfilariae (L1), taken up by a bloodmeal of the louse, develop into infective L3-stages, which are inoculated to seals during the blood meals [7,8,9]. The proposed role of E. horridus as an intermediate host for A. spirocauda was supported by the finding of different developmental stages in dissected lice [7, 9], the significant positive correlation of infestation of seals with E. horridus and seal infection with A. spirocauda [11], and molecular analyses [10].
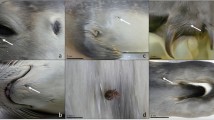
Echinophthiriid species exhibit special morphological characteristics, which are unique among anopluran lice and are the basis of their evolutionary success, being influenced by heterogeneous environmental conditions. Thus, distinct spiracles are equipped with special occlusions allowing for gas exchange [12]. Abdominal segments are characterised by a general loss of sclerotization and by a distinct trimming with modified setae [12, 13], sensorial organs, that are classified into spines, scales, and hairs [14]. Furthermore, the last two pairs of legs for fixation to the host surface are another characteristic of echinophthiriid lice. E. horridus differs from other members of the family by the restructuring of all three pairs of legs into claws and the absence of abdominal scale manifestation (Fig. 1) [14,15,17].
Due to these morphological adaptions and a strict specifity to semiaquatic host species, a coevolution of echinophthiriid lice and their hosts has been suggested [13, 18, 19]. Thereby, it has been proposed that the ancestors of pinnipeds must have already been infested with ancestral sucking lice before they ventured into the marine habitat [8, 20]. As a consequence of their growing specialisation to ancestral pinnipeds, ancestors of E. horridus lice might have evolved a vector role in the transmission of ancestors of the heartworm A. spirocauda, which is also believed to have undergone a coevolution with its host species [8]. Although filarial infections are usually transmitted by mosquitoes and ticks [21], this filarial heartworm nematode is believed to complete its life cycle in the seal louse E. horridus [7]. Nevertheless, it cannot be excluded that other intermediate hosts are involved in transmission of A. spirocauda, although suitable mosquito or tick vectors parasitising marine mammals were not reported.
Therefore, the present study aimed to investigate the possible role of E. horridus in transmission of pathogens, revealing new insights into the spectrum of potential vector-borne diseases circulating in free-ranging seals in the Dutch Wadden Sea.
Methods
Sample collection
During routine diagnostics at the Sealcentre Pieterburen (The Netherlands), sucking lice were sampled from 54 hospitalised harbour seals and from one grey seal admitted along the coast of the Dutch Wadden Sea (between 51°42′6.8″N, 3°40′42.1″ E and 53°27′57.6″N, 5°37′40.8″ E) between 6 May and 16 August 2012. Thereby, lice of harbour seals were collected opportunistically using lice combs and forceps and seals were not inspected thoroughly to minimise these potential stress factors. Care was taken to ensure that handling times were reduced to a minimum. If the lice infestation was severe, seals were treated with selamectin (topical) or ivermectin (subcutaneous). Regarding the age structure of the harbour seals, two animals were estimated at around 1 year of age, and all other seals were classified as weaners (< 6 months), including eight pups (< 4 weeks). Thirty of 54 harbour seals were males and 24 females. The grey seal examined in this study died despite intensive medical care during its rehabilitation period. In this case, lice were collected at necropsy.
Collected seal lice specimens as well as eggs (nits) from seal fur, mainly from the head and neck, were fixed in 70% ethanol and morphologically determined by light microscopy. All seal lice stages examined here were classified according to morphological features [1, 14,15,17]. All fixed specimens were kept at room temperature and delivered to the Institute of Parasitology, Justus Liebig University Giessen, Germany, for further processing. The estimation of the 95% CI was conducted using Kohn and Senyak [22].
PCR analyses
Seal lice were grouped from each seal and collection date separately, resulting in 64 pools from the 54 harbour seals (P. vitulina) containing 1–20 lice and one pool of > 1000 lice from the highly infested grey seal pup divided into pools of 15 lice (Additional file 1). DNA was extracted separately from each seal lice pool. Entire seal lice were washed in distilled water overnight and sliced to minute pieces with sterile scalpel blades in 100 µl of phosphate-buffered saline. DNA was extracted using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s instructions. PCR assays were performed to detect A. phagocytophilum, A. spirocauda, and Mycoplasma spp. DNA using as template 500 ng DNA extracted from the lice. For A. phagocytophilum detection, a partial gene sequence of the msp2 gene was amplified by real-time PCR according to Courtney et al. [23]. Mycoplasma was investigated by conventional PCR amplifying a partial 16S ribosomal DNA sequence with primers MGS0 and GP03 [24]. From the filarial species A. spirocauda, no sequences were available in NCBI GenBank at the time of this study (GenBank release 193, 12/2012). In a first attempt to obtain sequence data we used pan-filarial primers COIintF/COIintR [25] of the mitochondrial cytochrome c oxidase subunit 1 gene (cox1). The obtained sequences from five seal lice pools differed by only one nucleotide position and had a sequence identity of 90% to an Acanthocheilonema viteae cox1-sequence (HQ186249) in the GenBank database. The cox1 sequence from one seal lice pool was submitted to GenBank (accession number HG005138). To validate the sequences from the lice pools and to obtain further sequences from A. spirocauda, we then extracted DNA from morphologically confirmed A. spirocauda adult nematodes (identified according to Leidenberger and Boström [26]; please see Additional file 2), which were collected from necropsied harbour seals at Sealcentre Pieterburen between 2009-2011. Furthermore, we amplified, sequenced, and assembled a partial sequence of the ribosomal DNA region (ITS1, 5.8S, ITS2, and partial 28S) using primers NC5/NC2 [27] and the aforementioned cox1-primers.
PCR amplification products were separated on a 2% agarose gel and visualised using Midori Green. Sequencing was performed by an external service provider (LGC Genomics, Berlin, Germany) and the new sequences were deposited in GenBank database (accession numbers: A. spirocauda: HF583266, Mycoplasma sp.: MK953546). The cox1 amplicon sequence from the adult A. spirocauda (accession number HF583266) had an additional single-nucleotide polymorphism (not shown) compared to the sequences obtained from the DNA extracted from pools of infested lice.
For the molecular detection of A. spirocauda larvae in seal lice a more sensitive nested PCR (outer primers COIintF/COIintR; inner primers AspF/AspR) was established based on the alignment of cox1 sequences from A. spirocauda, A. viteae and Dirofilaria immitis. Primers AspF (5'-TGCTGTTACTTTGGACCAGGT-3') and AspR (5'-ATGATGGCCCCACACAGAAG-3') were designed with Primer3 software [28]. The first PCR (COIintF/COIintR) was performed in a reaction volume of 50 µl containing 5 µl template DNA, 5 µl 10× PCR buffer S (Peqlab, VWR International GmbH, Erlangen, Germany), 1 µl of each primer, 1 µl dNTPs, 1 µl 5 U/µl peqGOLD “Hot” Taq DNA Polymerase (Peqlab, VWR International GmbH, Erlangen, Germany), and 36 µl H20. The following conditions were used: 2 min 95 °C initial denaturation, 35 cycles of 30 s 94 °C, 30 s annealing at 50 °C, 45 s 72 °C, and 5 min 72 °C final extension. For the second PCR (AspF/AspR), 1 µl of the first PCR served as template in a 50 µl reaction containing 5 µl 10× PCR buffer S, 1 µl of each primer, 1 µl dNTPs, 1 µl peqGOLD “Hot” Taq DNA Polymerase, and 40 µl H20, under the following conditions: 2 min 95 °C initial denaturation, 35 cycles of 30 s 94 °C, 30 s annealing at 58 °C, 30 s 72 °C, and 5 min 72 °C final extension. PCR products were separated on a 2% agarose gel stained with Midori Green. The achieved sensitivity of this method was determined using the counted number of microfilariae (see Additional file 3) isolated from gravid A. spirocauda.
Phylogenetic analyses
For molecular phylogenetic analyses data sets of highly matching sequences to A. spirocauda and Mycoplasma sp. sequences were obtained from BLAST searches of the GenBank database (GenBank release 238, 06/2020); sequences were trimmed to homologous ends and realigned using the multiple sequence alignment program MAFFT 7 [29] with the L-INS-i method for the cox1 and 16S sequence data sets and the structure-aided Q-INS-i method for the ITS2 sequence data set. Phylogenetic trees were constructed using Bayesian analysis (MrBayes 3.2) (10.000 tree generations, sampling each 10, discarding first 250 trees) and TreeDyn for tree drawing at the phylogeny.fr platform [30]. The data sets included sequences homologous to nucleotides (nt) 122–624 of the A. spirocauda cox1 sequence (HF583266K), nt 466–942 of the A. spirocauda ITS2 sequence (HG005138), and nt 1–217 of the Mycoplasma sp. 16S ribosomal RNA gene sequence (MK953546). The A. spirocauda cox1 sequence of an isolate from Erignathus barbatus (bearded seal) in Barrow (Alaska, USA) (KF038155) [31] was not included because the overlapping sequence to the other A. spirocauda cox1 sequences (excluding KT899872) was too short (169 nt, intraspecies variation 2.4%, i.e. 4/169 nt).
Results
Seal lice and vector-borne pathogens
In total, 200 adult E. horridus seal lice were collected separately from 54 harbour seals (infestation 1–20 lice/seal, median 2; see Additional file 1) and more than 1000 lice from a single grey seal (H. grypus) pup. Seal lice from 19 of the 54 infested harbour seals (prevalence 35.2%; 95% CI 22.5–47.9%) and from the highly infested grey seal pup proved positive for A. spirocauda. In addition, seal lice (8 pools of 15 specimens) from the grey seal were found positive for A. phagocytophilum and seal lice from three harbour seals were found positive for Mycoplasma sp. Analysis of the partial 16S sequence revealed that a single strain of Mycoplasma sp. was present in the seal lice obtained from three harbour seals, which was identical to a Mycoplasma sp. from an Alaska native hunter who suffered from disseminated seal finger mycoplasmosis [32].
In an intial cox1-PCR using pan-filarial primers and sequencing the obtained sequences from lice pools and morphological confirmed A. spirocauda adults differed by only 3/649 nt (intraspecies variation 0.5%). We compared the cox1 single and nested PCR on ten pools. The nested PCR was more sensitive for these pools and therefore we decided to apply the nested PCR as diagnostic PCR for all pools. It had a consistent minimal detection of ten microfilariae (see Additional file 3) and was sufficient to detect A. spirocauda larvae in a single louse (seven positive lice pools from the harbour seals contained only one seal louse; see table in Additional file 1).
Phylogenetic analysis of A. spirocauda and the Mycoplasma isolate
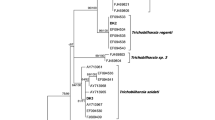
Phylogenetic analysis of the partial cox1 sequence showed that A. spirocauda was most closely related to A. odendhali (Fig. 2), a filarial parasite of the Northern fur seal and the California sea lion (Zalophus californianus) occurring in subcutaneous and intermuscular sites and considered to be non-pathogenic [31, 33]. Interestingly, one A. spirocauda isolate (PPr11-007; accession numbers cox1 KT899872, ITS2 MG581463) collected from a stranded, deceased harbour seal from the USA west coast (Stinson Beach, CA) [10] differs from all other A. spirocauda isolates by some indels in the ITS2 region and point mutations in the partial cox1 gene resulting in only 95% and 93% sequence identity respectively (not shown). The partial 16S sequence of the Mycoplasma species detected in seal lice of three harbour seals in this study was identical to the sequence from an infected human patient (GenBank accession no. KP292569). This zoonotic Mycoplasma species forms a new phylogenetic group together with Mycoplasma sp. isolates from common bottlenose dolphins (Tursiops truncatus) and from a California sea lion (Fig. 3). It is positioned next to the elephantis-equigenitalium group and clearly separated from other Mycoplasma species previously described infecting pinnipeds, such as M. phocicerebrale, M. phocidae, M. phocirhinis and M. zalophi.
Phylogenetic analysis of A. spirocauda. The phylogeny is based on the MAFFT alignments for a partial cox1 sequence and complete ITS2 rDNA region of A. spirocauda and other Acanthocheilonema species using Bayesian inference (Dirofilaria immitis sequence as outgroup). The A. dracunculoides ITS2 sequence is shorter and does not cover the whole sequence of A. spirocauda. Branch lengths are drawn proportionally to evolutionary distance (scale bar is shown). Numbers adjacent to nodes indicate posterior probabilities in per cent. Branch labels provide species names/host/two-letter country code/GenBank accession number
Phylogenetic analysis of Mycoplasma sp. from seal lice collected from harbour seals. Bayesian phylogenetic tree of Mycoplasma sp. of seal lice from harbour seals of the present study and phylogenetically closely related Mycoplasma species based on partial 16S gene sequences. Branch lengths are drawn proportionally to evolutionary distance (scale bar is shown). Numbers adjacent to nodes indicate posterior probabilities in per cent. Branch labels provide species names/host/source/two-letter country code/GenBank accession number. Mycoplasma species from seals highlighted in bold
Severe E. horridus infestation in a grey seal
Investigation of a grey seal which was found stranded on the beach revealed a severe E. horridus infestation (> 1000 specimens). The animal weighted 15 kg at admission to the Sealcentre Pieterburen and was estimated on its teeth development as a few week old pup. On arrival to the Sealcentre Pieterburen the male pup was cachectic, showing fever and severe dehydration. After 14 days of intensive medical care treatments, the pup did not improve and died.
At necropsy, gross pathology findings included consolidation of large portions of the lungs and presence of foam and fluids in the trachea and main bronchial trees. Histopathological examination of the lungs revealed acute interstitial pneumonia, which was diagnosed as the cause of animal death. No A. spirocauda infection was detected during necropsy. Unfortunately, no blood samples were left from this grey seal in order to perform blood smears and additional diagnostic PCRs to determine whether the grey seal was positive for A. phagocytophilum too.
Discussion
This study documents the presence of helminth and bacterial pathogens in the seal louse E. horridus, corroborating that this arthropod is a vector of A. spirocauda and suggesting that it may also play a role in the epidemiology of A. phagocytophilum and seal finger-associated Mycoplasma sp. The presence of A. spirocauda larvae in E. horridus was already described in the past [7,8,9, 10] and molecularly confirmed in the present study. Our results showing that seal lice from 35% of the harbour seals and from the grey seal proved positive for A. spirocauda strongly support the hypothesis that E. horridus functions as obligate intermediate host of A. spirocauda. Furthermore, A. spirocauda, the only filarial nematode parasite of phocid seals [8, 34], was molecularly characterised in this study.
Regarding the epidemiology and pathogenicity of echinophthiriosis, it is more frequently reported in young and weak animals [7, 8, 35], showing no seasonal variations for adult seals, but pups and immature seals have higher prevalences in spring. In contrast, Dailey and Fallace [11] reported the highest prevalence in the autumn and winter months, but no significant differences between examined age classes of seals and their seal lice burden were detected. Interestingly, in the closely related species A. microchir from the South American sea lion over 60% of 1-day-old pups were infested with lice, and recruitment increased in pups up to 3 days old and leveled off onwards. In 1-day-old pups, significantly more adults than nymphs were found, but this pattern was reversed in older pups, documenting the importance of vertical transmission probably through their mothers [36].
Molecular analyses on collected seal lice from harbour and grey seals revealed the presence of DNA of the seal heartworm A. spirocauda. In this context, E. horridus has previously been proposed to be the natural obligate intermediate host of A. spirocauda [7,8,9, 10], and different stages of A. spirocauda larvae were found in dissected E. horridus seal lice [7, 9]. So far, the heartworm A. spirocauda has been reported from different phocid species such as harbour seals, hooded seals (Cystophora cristata), bearded seals, ribbon seals (Phoca fasciata), harp seals (Phoca groenlandica), ringed seals, spotted seals (Phoca largha), monk seals (Monachus monachus), and recently from grey seals [8, 10, 37]. Furthermore, there is a significant positive correlation between heartworm infection and infestation of harbour seals with seal lice [11]. The nested PCR protocol developed in this study had a detection threshold as low as ten microfilariae and therefore may be a useful method for future studies on the epidemiology of these parasites in seal lice. Recently, Keroack et al. [10] developed a more sensitive A. spirocauda real-time quantitative PCR based on a highly repetitive genomic DNA repeat identified using whole-genome sequencing that could also improve future monitoring of seal heartworm infections. These authors also identified the first time an A. spirocauda adult worm in a presumed grey seal carcass from the coast of Cape Cod (MA, USA). However, the authors mentioned that the carcass was in very poor condition due to extensive decomposition and could not be fully identified and reported just the evidence for possible A. spirocauda infection in the grey seal.
Moreover, to our knowledge our results represent the first detection of A. phagocytophilum and Mycoplasma sp. in seal lice. However, based on the limited knowledge of vector-borne pathogens occurring in marine habitats, our findings of A. phagocytophilum and Mycoplasma sp. do not necessarily imply that the seal louse is a viable vector of these pathogens. Analogous to studies on the vector competence of human body lice [38, 39], evidence of vector competence of E. horridus and the ability of transmitting these pathogens could only be proved by experimental infections. Using molecular methods, it cannot be excluded that the pathogens detected were located in the gastrointestinal tract of E. horridus individuals after blood consumption of infected seals but are not able to be transmitted to other host individuals. Nevertheless, molecular surveys of the present study constitute important baseline studies in the field of marine mammal parasitology to initially reveal a spectrum of pathogens, which could possibly be transmitted by seal lice. Thereby, our results can also help to encourage other researchers to extend the knowledge of vector-borne pathogens in the field of marine mammal parasitology.
To our knowledge, there are no reports on A. phagocytophilum or Mycoplasma spp. in any other ectoparasite affecting marine mammals or evidence of anaplasmosis occurring in stranded phocid seals [40, 41]. Anaplasma phagocytophilum (Rickettsiales, Anaplasmataceae) constitutes an emerging globally distributed pathogen transmitted mainly by Ixodes ticks and may cause granulocytic anaplasmosis, one of the most relevant tick-borne diseases of veterinary and public health significance worldwide [42]. Virtually nothing is known about the occurrence and clinical implications of A. phagocytophilum in seals, and the detection of this pathogen in E. horridus is surprising since to our knowledge there are no records of tick parasitism on pinnipeds. This raises the question of whether other haematophagous ectoparasites (such as E. horridus) might play a role in the transmission of A. phagocytophilum among marine mammals.
Variable clinical signs of granulocytic anaplasmosis can include high fever, lethargy, inappetence, anorexia, dullness, reduced weight gain, coughing, and abortion in different animal species in Europe, including domestic ruminants, horses, dogs, and cats [42,43,44]. In humans, symptoms were reported as non-specific and include influenza-similar symptoms with fever and myalgia. In addition, leucopenia, thrombocytopenia and/or anaemia have been frequently reported to occur in certain A. phagocytophilum strain infections [43]. Many of these non-specific clinical signs were observed in the heavily E. horridus-infested grey seal pup evaluated in this study, for which ~ 50% of the seal louse pooled samples were positive for A. phagocytophilum, suggesting that this individual might have suffered from acute granulocytic anaplasmosis. The zoonotic potential of the A. phagocytophilum genetic variant detected here cannot be assessed by the msp2 sequence and needs further characterisation. In this context, it would also be interesting to investigate whether red foxes (Vulpes vulpes)—frequent hosts of A. phagocytophilum—living in the Dutch coastal dune area might occasionally feed on dead seal pups and get infested by E. horridus as has been reported for Arctic foxes (Vulpes lagopus) from Alaska and the fur seal louse (Antarctophthirus callorhini) [1]. In this context, it is worth noting that red fox activity was observed in the UK within a mainland grey seal breeding colony [45] and that satellite tracking studies have shown that grey seals captured in The Netherlands may migrate to breed in the UK [46].
Mycoplasma sp. was also molecularly identified in collected seal lice specimens in the current study, and in contrast to A. phagocytophilum there are previous reports on the occurrence of mycoplasmal infections in pinnipeds [47,48,49,50,51,52]. Mycoplasma spp. are common inhabitants of the respiratory, gastrointestinal and genital tracts of marine mammals, and a study on Australian fur seals (Arctocephalus pusillus doriferus) demonstrated the presence of different Mycoplasma species such as M. phocicerebrale, M. phocidae, M. zalophi, and Mycoplasma sp. in tested animals [49]. Furthermore, PCR testing of nasal swabs detected the presence of Mycoplasma spp. DNA in South American fur seal (Arctocephalus australis) populations in Peru with an estimated prevalence of around 38% [53], evidencing rather cosmopolitan distribution of mycoplasmas in pinnipeds. For universal detection of mycoplasmas the highly specific and sensitive PCR assay of van Kuppeveld [24] was used in the current study based on the conserved region of the 16S gene. Direct sequencing and sequencing of several clones indicated a single strain only. However, better species differentiation would be possible selecting further housekeeping genes (e.g. rpoB, rpoC) and culturing for phenotypical characterisation and serological testing. Therefore, our findings on Mycoplasma-positive E. horridus lice might suggest the presence of these bacterial infections in pinnipeds of the Dutch Wadden Sea. Whether seal lice might be potentially involved in the transmission of Mycoplasma will require further investigation.
Nevertheless, it is significant to note that some mycoplasmas (e.g. M. phocicerebrale) are associated with seal mortality and zoonotic ‘seal-finger’ infection, a disease known among people who handle seals [54]. Seal-finger lesions could progress to septic arthritis of the joints if tetracycline-based treatment is not received. Accordingly, more recent published studies on a series of bites and contact abrasion in open-water swimmers caused by California sea lions and harbour seals revealed the presence of Mycoplasma spp. in human wounds [52, 55], demonstrating its zoonotic potential. GenBank database search using the partial Mycoplasma sp. 16S sequence, detected in seal lice collected from three harbour seals in the current study resulted in 100% identity to the sequence of an unnamed Mycoplasma strain (GenBank accession number KP292569) obtained from a patient with “seal finger” and an infected hip joint. The patient previously hunted and harvested ringed seals (Phoca hispida) in Alaska without protective gloves in an area where Mycoplasma-infected seals had been noticed before [32].
Conclusions
In conclusion, this study shows the occurrence of potential pathogens within seal lice of infested pinnipeds in the Dutch Wadden Sea and reveals new insights into the potential role of E. horridus as vector. Thereby, the current study molecularly confirms former suggestions that E. horridus functions as an obligate intermediate host for the seal heartworm A. spirocauda. Whether E. horridus plays a role in the transmission of granulocytic anaplasmosis and Mycoplasma spp. infections in seals remains to be clarified by future studies.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Leonardi MS, Palma RL. Review of the systematics, biology and ecology of lice from pinnipeds and river otters (Insecta: Phthiraptera: Anoplura: Echinophthiriidae). Zootaxa. 2013;3630:445–66.
Leonardi MS, Poljak S, Carlini P, Galliari J, Bobinac M, Santos M, et al. Antarctophthirus carlinii (Anoplura: Echinophthiriidae), a new species from the Weddell seal Leptonychotes weddelli. Parasitol Res. 2014;113:3947–51.
Jellison WL, Milner KC. Salmonellosis (bacillary dysentery) of fur seals. J Wildl Manag. 1958;22:199–200.
Boyd BM, Allen JM, Koga R, Fukatsu T, Sweet AD, Johnson KP, et al. Two bacterial genera, Sodalis and Rickettsia, associated with the seal louse Proechinophthirus fluctus (Phthiraptera: Anoplura). Appl Environ Microbiol. 2016;82:3185–97.
Morick D, Osinga N, Gruys E, Harrus S. Identification of a Bartonella species in the harbor seal (Phoca vitulina) and in seal lice (Echinophthirius horridus). Vec Born Zoo Dis. 2009;9:751–3.
Linn ML, Gardner J, Warrilow D, Darnell GA, McMahon CR, Field I, et al. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J Virol. 2001;75:4103–9.
Geraci JR, Fortin JF, St Aubin DJ, Hicks BD. The seal louse, Echinophthirius horridus: an intermediate host of the seal heartworm, Dipetalonema spirocauda (Nematoda). Can J Zool. 1981;59:1457–9.
Leidenberger S, Harding K, Härkönen T. Phocid seals, seal lice and heartworms: a terrestrial host-parasite system conveyed to the marine environment. Dis Aquat Organ. 2007;77:235–53.
Lehnert K, Schwanke E, Hahn K, Wohlsein P, Siebert U. Heartworm (Acanthocheilonema spirocauda) and seal louse (Echinophthirius horridus) infections in harbour seals (Phoca vitulina) from the North and Baltic Seas. J Sea Res. 2016;113:65–72.
Keroack CD, Williams KM, Fessler MK, DeAngelis KE, Tsekitsidou E, Tozloski JM, et al. A novel quantitative real-time PCR diagnostic assay for seal heartworm (Acanthocheilonema spirocauda) provides evidence for possible infection in the grey seal (Halichoerus grypus). Int J Parasitol Parasites Wildl. 2018;7:147–54.
Dailey MD, Fallace LS. Prevalences of parasites in a wild population of the Pacific harbour seal (Phoca vitulina richardsi) from Gray’s Harbor, Washington. Bull South Calif Acad Sci. 1989;88:1–10.
Kim KC. Ecological and morphological adaption of the sucking lice (Anoplura, Echinophthiriidae) on the Northern fur seal. Rap Proces. 1975;169:504–15.
Kim KC. The sucking lice (Anoplura: Echinophthiriidae) of the Northern fur seal; descriptions and morphological adaptations. Ann Entomol Soc Am. 1971;64:280–92.
Mehlhorn B, Mehlhorn H, Plötz J. Light and scanning electron microscopical study on Antarctophthirus ogmorhini lice from the Antarctic seal Leptonychotes weddellii. Parasitol Res. 2002;88:651–60.
Scherf H. Ein Beitrag zur Kenntnis zweier Pinnipedierläuse (Antarctophthirus trichechi Boheman und Echinophthirius horridus Olfers). Z Parasitenk. 1963;23:16–44.
Miller FH Jr. Scanning electron microscopy of Echinophthirius horridus (von Olfers), Antarctophthirus callorhini (Osborn), and Proechinophthirius fluctus (Ferris) with emphasis on the antennal structures (Anoplura: Echinophthiriidae). J Parasitol. 1971;57:668–74.
Beder G. Rasterelektronenmikroskopische Studie der Robbenlaus Echinophthirius horridus (Olfers 1816). M D Gesell Allg Ange. 1990;7:512–6.
Kim KC. Coevolution of parasitic arthropods and mammals. New York: Wiley; 1985.
Leonardi MS, Virrueta Herrera S, Sweet A, Negrete J, Johnson KP. Phylogenomic analysis of seal lice reveals codivergence with their hosts. Syst Entomol. 2019;44:699–708.
Kim KC, Repenning CA, Morejohn GV. Specific antiquity of the sucking lice and evolution of otariid seals. Rap Proces. 1975;169:544–9.
Lefoulon E, Bain O, Bourret J, Junker K, Guerrero R, Cañizales I, et al. Shaking the tree: multi-locus sequence typing usurps current onchocercid (Filarial Nematode) phylogeny. PLoS Negl Trop Dis. 2015;9:e0004233.
Kohn MA, Senyak J. Sample size calculators. 2020. https://www.sample-size.net/. Accessed 27 Nov 2020.
Courtney JW, Kostelnik LM, Zeidner NS, Massung RF. Multiplex real-time PCR for detection of Anaplasma phagocytophilum and Borrelia burgdorferi. J Clin Microbiol. 2004;42:3164–8.
van Kuppeveld FJM, Melchers WJG, Willemse HFM, Kissing J, Galama JMD, van der Logt JTM. Detection of Mycoplasma pulmonis experimentally infected laboratory rats by 16S rRNA amplification. J Clin Microbiol. 1993;31:524–7.
Casiraghi M, Anderson TJC, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122:93–103.
Leidenberger S, Boström S. Characterization of the heartworm Acanthocheilonema spirocauda (Leidy, 1858) Anderson, 1992 (Nematoda: Onchocercidae) in Scandinavia. Parasitol Res. 2008;104:63–7.
Gasser RB, Hoste H. Genetic markers for closely-related parasitic nematodes. Mol Cell Probe. 1995;9:315–9.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucl Acids Res. 2012;40:e115.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res. 2008;36:465–9.
Kuzmina TA, Kuzmin YI, Tkach VV, Spraker TR, Lyons ET. Ecological, morphological, and molecular studies of Acanthocheilonema odendhali (Nematoda: Filarioidea) in northern fur seals (Callorhinus ursinus) on St. Paul Island, Alaska. Parasitol Res. 2013;112:3091–100.
Westley BP, Horazdovsky RD, Michaels DL, Brown DR. Identification of a novel Mycoplasma species in a patient with septic arthritis of the hip and seal finger. Clin Infect Dis. 2016;62:491–3.
Farriols M, Arellano-Carbajal F, Elorriaga-Verplancken FR, Adame-Fernández K, Garrido E, Álvarez-Martínez RC, et al. Filarial infections in California sea lions vary spatially within the Gulf of California, Mexico. Parasitol Res. 2020;119:1281–90.
Lehnert K, Raga JA, Siebert U. Parasites in harbour seals (Phoca vitulina) from the German wadden sea between two phocine distemper virus epidemics. Helgol Mar Res. 2007;61:239–45.
Thompson PM, Corpe HM, Reid RJ. Prevalence and intensity of the ectoparasite Echinophthirius horridus on harbour seals (Phoca vitulina): effects of host age and inter-annual variability in host food availability. Parasitology. 1998;117:393–403.
Leonardi MS, Crespo EA, Raga JA, Aznar FJ. Lousy mums: patterns of vertical transmission of an amphibious louse. Parasitol Res. 2013;112:3315–23.
Papadopoulos E, Loukopoulos P, Komnenou A, Androukaki E, Karamanlidis AA. First report of Acanthocheilonema spirocauda in the Mediterranean monk seal (Monachus monachus). J Wildl Dis. 2010;46:570–3.
Houhamdi L, Lepidi H, Drancourt M, Raoult D. Experimental model to evaluate the human body louse as a vector of plague. J Infect Dis. 2006;194:1589–96.
Houhamdi L, Fournier PE, Fang R, Lepidi H, Raoult D. An experimental model of human body louse infection with Rickettsia prowazekii. J Infetc Dis. 2002;186:1639–46.
Sonne C, Lakemeyer J, Desforges JP, Eulaers I, Persson S, Stokholm I, et al. A review of pathogens in selected Baltic Sea indicator species. Environ Int. 2020;137:105565.
Colegrove KM, Burek-Huntington KA, Roe W, Siebert U. Pinnipediae. In: Terio KA, McAloose D, St. Leger J, editors. Pathology of Wildlife and Zoo Animals. Cambridge: Academic Press; 2008. p. 569–92.
Atif FA. Anaplasma marginale and Anaplasma phagocytophilum: Rickettsiales pathogens of veterinary and public health significance. Parasitol Res. 2015;114:3941–57.
Dugat T, Lagrée AC, Maillard R, Boulouis HJ, Haddad N. Opening the black box of Anaplasma phagocytophilum diversity: Current situation and future perspectives. Front Cell Infect Microbiol. 2015;5:61.
Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum—a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Mi. 2013;3:31.
Culloch RM, Pomeroy P, Lidstone-Scott R, Bourne L, Twiss SD. Observations from video footage of red fox (Vulpes vulpes) activity within a grey seal (Halichoerus grypus) breeding colony on the UK mainland. Aquat Mamm. 2012;38:81–5.
Brasseur S, Aarts G, Kirkwood R. Habitat quality for Grey Seals in the Dutch Wadden Sea.. IMARES Wageningen UR 2015; Report number C090-14.
Ruhnke HL, Madoff S. Mycoplasma phocidae sp. nov, isolated from harbor seals (Phoca vitulina L.). Int J Syst Evol Micr. 1992;42:211–4.
Haulena M, Gulland FMD, Lawrence JA, Fauquier DA, Jang S, Aldridge B, et al. Lesions associated with a novel Mycoplasma sp. in California sea lions (Zalophus californianus) undergoing rehabilitation. J Wildl Dis. 2006;42:40–5.
Lynch M, Taylor TK, Duignan PJ, Swingler J, Marenda M, Arnould JP, et al. Mycoplasmas in Australian fur seals (Arctocephalus pusillus doriferus): identification and association with abortion. J Vet Diagn Invest. 2011;23:1123–30.
Volokhov DV, Norris T, Rios C, Davidson MK, Messick JB, Gulland FM, et al. Novel hemotrophic mycoplasma identified in naturally infected California sea lions (Zalophus californianus). Vet Microbiol. 2011;149:262–8.
Tryland M, Nesbakken T, Robertson L, Grahek-Ogden D, Lunestad BT. Human pathogens in marine mammal meat–a northern perspective. Zoonoses Public Health. 2014;61:377–94.
Nuckton TJ, Simeone CA, Phelps RT. California sea lion (Zalophus californianus) and harbor seal (Phoca vitulina) bites and contact abrasions in open-water swimmers: a series of 11 cases. Wilderness Environ Med. 2015;26:497–508.
Jankowski G, Adkesson MJ, Saliki JT, Cárdenas-Alayza S, Majluf P. Survey for infectious disease in the South American fur seal (Arctocephalus australis) population at Punta San Juan, Peru. J Zoo Wildl Med. 2015;46:246–54.
White CP, Jewer DD. Seal finger: a case report and review of the literature. Can J Plastic Surg. 2009;17:133–5.
Kornblith AE, Budak JZ, Simeone CA, Nuckton TJ. Severe sea lion bites in urban cold-water swimmers. J Emerg Med. 2019;57:859–65.
Acknowledgements
We kindly acknowledge the animal care staff and all international volunteers of Sealcentre Pieterburen for helping us in sample collection as well as Christine Henrich (Institute of Parasitology, JLU Giessen) for her excellent assistance in molecular characterization of pathogens in seal lice.
Funding
Open Access funding enabled and organized by Projekt DEAL.. Open Access funding enabled and organized by Projekt DEAL. This study was funded by the Institute of Parasitology (Justus Liebig University Giessen) and the Sealcentre Pieterburen (The Netherlands).
Author information
Authors and Affiliations
Contributions
CH designed the study. CH, JH and DE drafted the manuscript. JH conducted molecular analyses of lice samples and A. spirocauda. JH and DE contributed equally to this work. GSC and ARG provided support during sample taking at Sealcentre Pieterburen. GM and UG provided scanning electron microscopic images. AT essentially revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All parasite specimens analyzed in the current study (E. horridus and adult A. spirocauda individuals) were exclusively provided by the Sealcentre Pieterburen, The Netherlands, sampled from free-ranging harbour and grey seals during a rehabilitation period or necropsies. Seal sampling procedures were conducted in strict accordance to the permission of the government of The Netherlands (valid permission ID at time of study: FF/75/2012/015), which approves admission and rehabilitation of free-ranging seals at the Sealcentre Pieterburen as well as collection of dead seals. In the present study, a non-invasive lice sample-taking process (using lice combs) was conducted during routine veterinary diagnostics and therapy to minimise additional stress factors and sampling duration. No seals were euthanized for research purposes. All samples of the present study were exclusively taken on territory of The Netherlands. The Sealcentre Pieterburen granted permission to the Institute of Parasitology (Justus Liebig University Giessen) for further analyses of parasite samples to perform the present study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Infestation of individual seals with seal lice.
Additional file 2.
Morphological features of A. spirocauda.
Additional file 3.
Sensitivity of the Acanthocheilonema spirocauda cox1 nested-PCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hirzmann, J., Ebmer, D., Sánchez-Contreras, G.J. et al. The seal louse (Echinophthirius horridus) in the Dutch Wadden Sea: investigation of vector-borne pathogens. Parasites Vectors 14, 96 (2021). https://doi.org/10.1186/s13071-021-04586-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-04586-9