Abstract
Background
The African leopard Panthera pardus pardus (L.) is currently listed as a vulnerable species on the IUCN (International Union for the Conservation of Nature) red list of threatened species due to ongoing population declines. This implies that leopard-specific parasites are also vulnerable to extinction. Intracellular apicomplexan haemoparasites from the genus Hepatozoon Miller, 1908 have been widely reported from wild carnivores in Africa, including non-specific reports from leopards. This paper describes two new haemogregarines in captive and wild leopards from South Africa and provides a tabular summary of these species in relation to species of Hepatozoon reported from mammalian carnivores.
Methods
Blood was collected from nine captive and eight wild leopards at various localities throughout South Africa. Thin blood smears were Giemsa-stained and screened for intraleukocytic haemoparasites. Gamont stages were micrographed and morphometrically compared with existing literature pertaining to infections in felid hosts. Haemogregarine specific primer set 4558F and 2733R was used to target the 18S rRNA gene for molecular analysis. Resulting sequences were compared to each other and with other available representative mammalian carnivore Hepatozoon sequences from GenBank.
Results
Two species of Hepatozoon were found in captive and wild leopards. Of the 17 leopards screened, eight were infected with one or both morphologically and genetically distinct haemogregarines. When compared with other species of Hepatozoon reported from felids, the two species from this study were morphometrically and molecularly distinct. Species of Hepatozoon from this study were observed to exclusively parasitize a particular type of leukocyte, with Hepatozoon luiperdjie n. sp. infecting neutrophils and Hepatozoon ingwe n. sp. infecting lymphocytes. Phylogenetic analysis showed that these haemogregarines are genetically distinct, with Hepatozoon luiperdjie n. sp. and Hepatozoon ingwe n. sp. falling in well supported separate clades.
Conclusions
To our knowledge, this is the first morphometric and molecular description of Hepatozoon in captive and wild African leopards in South Africa. This study highlights the value of using both morphometric and molecular characteristics when describing species of Hepatozoon from felid hosts.
Similar content being viewed by others
Background
Members of the genus Hepatozoon Miller, 1908 are intracellular apicomplexan haemogregarines (Apicomplexa Levine, 1970: Adeleorina Léger, 1911: Hepatozoidae Wenyon, 1926) widely reported from amphibians, reptiles, birds and mammals, specifically including carnivores such as wild felids [1]. Feline hepatozoonosis was first reported in the early 1900’s by Patton [2], who described an intraleukocytic parasite Leucocytozoon felis domestici Patton, 1908 from an domestic cat in India. Eighteen years later, Wenyon [3] distinguished a capsule encasing this parasite that was overlooked by Patton [2], and then reclassified this parasite as a species of Hepatozoon, apparently morphologically identical to those found in hyenas, dogs and jackals [2, 3]. In the late 1990’s Beaufils et al. [4] proposed that canid and felid Hepatozoon are separate species, due to their morphological differences. The majority of species of Hepatozoon detected in African carnivores were previously identified as Hepatozoon canis (James, 1905), until Levine [5] suggested that host species should be taken into account when determining the identity of these parasites. Peirce et al. [6] supported this by suggesting that researchers are wrong to assume that all species of Hepatozoon from African carnivore are synonyms of H. canis.
From a parasitological perspective, the leopard Panthera pardus (L.) is one of the least studied big cats. Known protozoan parasites in leopards include species of Toxoplasma (Nicolle & Manceaux, 1908), Sarcocystis Lankester, 1882, Hepatozoon, Giardia Künstler, 1882 and Isospora Schneider, 1881 [7]. Brocklesby & Vidler [8] were the first to report Hepatozoon-like organisms from a free-ranging African leopard P. pardus pardus (Linnaeus 1758) in Kenya. In the early 1970’s, Keymer [9] reported Hepatozoon canis-like schizonts from the cardiac muscle of a leopard in Central Africa and four years later McCully et al. [10] reported Hepatozoon-like parasites from leopards in South Africa’s KwaZulu-Natal Province. More recently, Pawar et al. [11] and Khoshnegah et al. [12] detected what they thought to be Hepatozoon felis (Patton, 1908) from two free-ranging Indian leopards Panthera pardus fusca (Meyer) as well as an unnamed species of Hepatozoon from a single free-ranging Persian leopard Panthera pardus ciscausica (Satunin).
Prior to the 21st century, classification of species of Hepatozoon was based on their life history, host identity and morphological characteristics. However, with recent advances in molecular techniques, phylogenetic analyses on the relationships between species have become possible [13,14,15,16]. Morphological characteristics used to distinguish between species of Hepatozoon include gamont and nucleus dimensions; position of the nucleus within the gamont; number and arrangement of vacuoles and staining properties [17, 18], as well as characteristics of other developmental stages [19, 20]. In Africa, domestic dogs and wild carnivores have been reported to have cases of asymptomatic hepatozoonosis, caused mostly by H. canis and H. felis [6, 10, 21,22,23,24,25]. Although, H. canis and H. felis are not specific to either canids or felids, they have been reported infecting both carnivore families [26,27,28].
Due to constant improvement of molecular techniques, the number of studies on haemogregarines has systematically increased, with several studies relying solely on these methods to detect species of Hepatozoon in their hosts [11, 28,29,30,31,32]. Recent research has shown that a difference in p-distance of between 1–2% of the 18S rRNA gene is sufficient to distinguish between species of haemogregarines if supported by morphological data [16, 18, 33, 34]. Other studies, such as Metzger et al. [35], incorporate both molecular and some degree of morphological investigations, showing the importance of utilizing a more holistic approach when distinguishing among species of Hepatozoon.
The objectives of the present study were to (i) investigate whether captive and wild leopards in South Africa are infected with species of Hepatozoon; (ii) identify any infections found using both molecular analysis of a fragment of the 18S rRNA gene and morphological characteristics of the gamont stage in peripheral blood; (iii) determine if any of the Hepatozoon spp. identified can be linked to hepatozoonosis based on clinical symptoms of the host. This study is the first report on the molecular and morphological characteristics of Hepatozoon species infecting captive and wild leopards in South Africa.
Methods
Study area, Panthera pardus pardus collection and blood preparation
Blood samples were obtained from nine captive and eight wild leopards in South Africa (Table 1), representative of three core wild populations as identified by Daly et al. [36]. All live leopards were sedated and sampled by a qualified veterinarian surgeon following standard procedures. Blood scabs were collected from the frozen carcass of an erythristic (red) female leopard in the Mpumalanga Province (25°9′51.31ʺS, 30°26′55.46ʺE). Captive-bred leopards were sampled at two facilities in Bloemfontein, Free State Province (29°4′9.32ʺS, 26°9′34.40ʺE) (29°6′52.28ʺS, 26°12′15.80ʺE), and at one facility in the Mpumalanga Province (24°30′52.44ʺS, 30°54′8.82ʺE). Wild leopards were sampled in the Mpumalanga (25°9′51.31ʺS, 30°26′55.46ʺE) (24°34′43.50ʺS, 31°25′46.48ʺE) and northern Limpopo provinces (23°02′17.1ʺS, 29°26′26.5ʺE). Data collected on leopards included: geographical location, weight, age, physical condition, diet, parasite treatment, possible injuries, physical measurements, and possible clinical symptoms.
For this study, leopards are classified as ‘captive’ if they were born or raised in captivity from an early age (< 1-month-old). Leopards are classified as ‘wild’ when born or caught at an adult stage in the wild or when held at rehabilitation centres for less than six months to be relocated. Age classes were allocated to all leopards sampled following Fattebert et al. [37], with cubs (< 1 year), subadults (1–3 years) and adults (> 3 years). Wild leopards were aged according to the nature of tooth wear and a combination of morphological cues as described by Stander [38] and Balme et al. [39] respectively. Captive leopards were aged according to the information provided by their ex situ managers.
Prior to blood collection, the dense pelt over the identified area of collection was shaved with a clipper and wiped with an alcohol-soaked cotton ball, which helped remove external skin contaminants and improved visualisation of the vein. Peripheral blood was usually collected from the jugular or the cephalic vein by venipuncture with the use of a sterile Vacutainer system, in BD Vacutainer® (Franklin Lakes, USA) CAT (Clot Activator Tubes) and Vacutainer® EDTA tubes for molecular analysis.
Small blood droplets (enough to provide three to four duplicate blood smears) were placed onto clean, pre-labelled microscope slides to make thin blood smears. Blood smears were air-dried and subsequently fixed with absolute methanol for one minute. Once dry, blood smears were stored in slide boxes for further processing in the lab. A modified Giemsa (Fluka, Sigma-Aldrich, Steinheim, Germany) stain solution was prepared with distilled water (ratio of 9:1) in a 50 ml staining container. Air-dried blood smears were stained in the Giemsa solution for 20 min, rinsed with a slow stream of distilled water and again left to air dry.
Screening of blood smears
Stained smears were examined under the 100× oil immersion objective of a Nikon Eclipse E800 compound microscope (Nikon, Amsterdam, The Netherlands) and digital images of any infections detected were captured with an attached Nikon DS-Fi1 digital camera and accompanying software. Haemoparasites were identified through comparison of morphometric data to previous studies on species of Hepatozoon from carnivores [12, 19]. Parasitaemia was calculated per 100 host cells, with ~ 500 host cells (ten fields of 50 host cells) examined per blood smear. Photomicrographs of blood smears were calibrated according to the guidelines stipulated by the ImageJ Image Processing and Analysis software [40]. Measurements of parasites and leopard blood cells were taken with the ImageJ version 1.47 software program (Wayne Rasband National Industries of Health, USA) (http://imagej.nih.gov/ij). All measurements are in micrometres and are given as the range followed by the mean ± standard deviation (SD) in parentheses.
Molecular analysis
Blood samples collected directly into Vacutainer® EDTA tubes were thawed and used for molecular protocols. DNA was extracted with the KAPA Blood PCR Kit B (Kapa Biosystems, Cape Town, South Africa) according to the protocol provided by the manufacturer. DNA for the dried blood collected from the erythristic leopard was extracted by using the Kapa Express DNA extraction kit (Kapa Biosystems, Cape Town, South Africa), following the manufacturer’s protocols.
Haemogregarine-specific primers 4558F (5ʹ-GCT AAT ACA TGA GCA AAA TCT CAA-3ʹ) and 2733R (5ʹ-CGG AAT TAA CCA GAC AAA T-3ʹ) [41] were used for the detection of Hepatozoon species through PCR (polymerase chain reaction) amplification of the 18S rRNA gene. Fragments of between 995 and 1002 nucleotide (nt) were amplified using the primer set as mentioned above. PCRs were performed with 1.25 µl (10 µM) of each primer, 12.5 µl Kapa Blood Mix B, 7.5 µl molecular grade nuclease-free water (Thermo Fisher Scientific, Vilnius, Lithuania) and 2.5 µl whole blood to make up a final volume of 25 µl per sample. The PCR was undertaken in a Bio-Rad C1000 TouchTM Thermal Cycler PCR machine (Bio-Rad, Hemel Hempstead, UK), under the following conditions. Initial denaturation step of 5 min at 95 °C, followed by 35 cycles of denaturation for 30 s at 95 °C, annealing for 30 s at 50 °C and extension for 1 min at 72 °C. This was followed by a final extension of 7 min at 72 °C, and products were held at 4 °C. Resulting amplicons were visualised on a 1% agarose gel stained with gel red and using a Bio-Rad Gel-DocTM XR+ imaging system (Bio-Rad, Hemel Hempstead, UK) under ultraviolet light.
All positive, purified PCR products were sent for sequencing to Inqaba Biotechnical Industries (Pty) Ltd. (IBSA) (Pretoria, South Africa), a commercial sequencing company, for sequencing in both directions. Resultant sequences species identity was verified against previously published sequences using the Basic Local Alignment Search Tool (BLAST) [42]. Haemogregarine species identity was determined by establishing the closest BLAST match (97–100% to existing sequences available on the GenBank database). All sequences matching Hepatozoon spp. were considered positive and, since they were identical within each new species, only one representative sequence of each was included in further analysis.
The software package Geneious R11 (http://www.geneious.com [43]) was used to assemble and edit resultant sequence fragments. Sequences were aligned using the Clustal W 2.1 alignment tool [44] implemented within Geneious R11. A model test was performed using jModelTest 2·1·7 [45], to determine the most suitable nucleotide substitution model, according to the Bayesian information criterion (BIC). The model with the best BIC score was the General Time Reversible [46] model with estimates of invariable sites and a discrete Gamma distribution (GTR+I+G). 18S rDNA sequences for species of Hemolivia Petit, Landau, Baccam & Lainson, 1990, Hepatozoon Miller, 1908, Karyolysus Labbé, 1894, Haemogregarina Danilewsky, 1885 and Dactylosoma Labbé, 1894 (parasitising amphibian, reptilian and mammalian hosts) were downloaded from GenBank and aligned with the sequences generated in this study (Table 2). Adelina dimidiate Schneider, 1875, Adelina grylli Butaeva, 1996 (GenBank: DQ096835-DQ096836) and Klossiella equi Smith & Johnson, 1902 (GenBank: MH211602), from the suborder Adeleiorina Léger, 1911 were selected as outgroup. Although eight sequences were obtained from infected leopards, only a single representative of each species of Hepatozoon amplified in the present study was used for phylogenetic analyses. Phylogenetic analyses consisted of two datasets, the first alignment a large dataset (n = 297) including all representative H. felis sequences from GenBank (Additional file 1: Figure S1), and the second alignment based on the results of the first included 57 representative sequences (Table 2). Bayesian inference (BI) was used to infer phylogenetic relationships. The BI analysis was performed using MrBayes 3.2.2 [47] implemented from within Geneious R11. To assess posterior probability support the Markov Chain Monte Carlo (MCMC) algorithm was run for one million generation for the first larger dataset (297 sequences), and 10 million generations for the second the smaller dataset (57 sequences), sampling every 100 generations and using the default parameters. The first 25% of the trees were discarded as ‘burn-in’ with no ‘burn-in’ samples being retained. Results were visualised in Trace, to assess convergence and the ‛burn-inʼ period. Furthermore, uncorrected p-distances for the sequences used were also calculated in PAUP (Phylogenetic Analysis Using Parsimony) version 4.0a152 (Additional file 2: Table S1).
Results
Taxonomy
Phylum Apicomplexa Levine, 1970
Class Conoidasida Levine, 1988
Order Eucoccidiorida Léger & Dubosq, 1910
Suborder Adeleorina Léger, 1911
Family Hepatozoidae Wenyon, 1926
Genus Hepatozoon Miller, 1908
Hepatozoon luiperdjie n. sp.
Type-host:Panthera pardus pardus (L.) (Carnivora: Felidae).
Type-locality: Lajuma Research Centre (23°02ʹ17.1ʺS, 29°26ʹ26.5ʺE), Limpopo Province, South Africa.
Other localities: Greater Kruger Conservation Area (24°34ʹ43.50ʺS, 31°25ʹ46.48ʺE), Mpumalanga Province; Lydenburg area (25°9ʹ51.31ʺS, 30°26ʹ55.46ʺE), Mpumalanga Province, South Africa.
Type-material: Hapantotype, 1 peripheral blood smear from the type-host P. p. pardus and type-locality (23°02ʹ17.1ʺS, 29°26ʹ26.5ʺE), deposited under the accession number NMBP392 in the protozoan collection of the National Museum, Bloemfontein, South Africa.
Vector: Unknown.
Representative DNA sequences: Two sequences, of a 995 nt fragment of the 18S rRNA gene of Hepatozoon luiperdjie n. sp., isolated from the type-host P. p. pardus, deposited under the accession numbers MN793002 and MN793003 in the GenBank database.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [48], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:9E65A924-729F-43A5-AE1F-8001204B6A6A. The LSID for the new name Hepatozoon luiperdjie n. sp. is urn:lsid:zoobank.org:act:2293B0B3-3B91-4BDC-8246-50215B80F8D5.
Etymology: The species epithet is derived from the Afrikaans language diminutive name for the host P. p. pardus, which in Afrikaans is referred to as “luiperdjie”.
Description
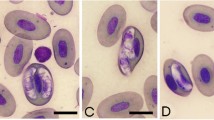
Gamonts. Most abundant stage in peripheral blood smears (Fig. 1a-f). Extracellular forms (Fig. 1c) and immature gamonts (Fig. 1d) rarely observed and no division stages detected. Mature gamonts measure 9.9–12.6 × 4.1–5.0 (11.0 ± 0.9 × 4.7 ± 0.4) (n = 53), area of 39.5–46.2 (42.0 ± 2.9) µm2 (n = 53). Mature gamonts mostly conspicuous within neutrophil cytoplasm (Fig. 1a–c), sometimes hardly visible and concealed by host cell nucleus; elongate, with bluntly rounded extremities, thin visible capsule (Fig. 1a); cytoplasm stained pale purple with some gamonts containing bright magenta and basophilic staining granules (Fig. 1a, f). Some gamonts with 2 to 3 small, slightly noticeable posteriorly situated vacuoles (Fig. 1e, thick arrow). Gamont nuclei measure 3.0–3.6 × 3.1–3.8 (3.5 ± 0.3 × 3.4 ± 0.3) (n = 53), area of 8.8–9.6 (9.2 ± 0.3) µm2 (n = 53); rounded and acentric, usually as wide as gamont at widest point, mostly located closer to anterior than posterior of gamont and stained dark purple, with densely stranded chromatin. Capsules 0.3–0.7 (0.5 ± 0.2) thick (n = 53) and observable in most gamonts (Fig. 1e, thin arrow).
a–f Peripheral blood gamont stages of Hepatozoon luiperdjie n. sp. in the African leopard Panthera pardus pardus from hapantotype slide (NMBP392). a, b, e, f Mature gamonts within neutrophils, where enlargement of host cell and displacement of host cell nucleus is apparent. c Extracellular gamont. d Immature gamont. e Mature gamont in which small posterior vacuoles (thick arrow) and thin capsule (thin arrow) can be seen. c, f Disintegration of neutrophils by infecting gamonts. g–k Peripheral blood gamont stages of Hepatozoon ingwe n. sp. in the African leopard Panthera pardus pardus from hapantotype slide (NMBP393). g, h, k Mature gamonts within lymphocytes, where lateral compression of host cell is apparent. g Mature gamont in which bright pink granules and thin capsule (thick arrow) can be seen. h Mature gamont with prominent posterior vacuoles (thin arrow). i Extracellular gamont. j Immature gamont. k Co-infection of Hepatozoon luiperdjie n. sp. (on the left) and Hepatozoon ingwe n. sp. in the same leopard. Scale-bar: 10 µm
Prevalence and parasitaemia.Hepatozoon luiperdjie n. sp. occurred in peripheral blood of 8/16 (prevalence 50%) individual P. p. pardus sampled. This haemogregarine formed co-infections with Hepatozoon ingwe n. sp. (see below) in 6 out of 16 leopards (prevalence of 38%), and was the sole species of Hepatozoon detected in 2 out of 16 individuals (prevalence of 13%). Prevalence of 63% (5/8) in males and 38% (3/8) in females infected. No captive individuals infected by this haemogregarine. Parasitaemia varied between individuals and could only be determined in WM1 (11.3%), WF1 (21.5%), WM3 (1.9%), WF3 (15.4%) and WM4 (4.8%). Average parasitaemia in all five leopards was 11.0%.
Effect on host cells. Gamonts sometimes compressed the lobulated nucleus of neutrophils, either towards the periphery of the host cell (Fig. 1a), or towards one side (Fig. 1b). Parasitized neutrophils measured 12.7–14.3 × 12.1–14.1 (13.6 ± 0.8 × 13.1 ± 0.7) (n = 53), area of 130.2–186.0 (146.3 ± 22.9) µm2 (n = 53). Healthy, uninfected neutrophils measured 7.6–14.4 × 7.0–13.0 (11.1 ± 1.5 × 10.3 ± 1.3) (n = 450), area of 46.3–127.2 (89.1 ± 18.3) µm2 (n = 450). Nuclei of parasitized neutrophils measured 12.8–25.3 × 3.3–13.3 (19.8 ± 5.2 × 5.7 ± 4.2) (n = 53), area of 53.3–116.9 (68.8 ± 27.0) µm2 (n = 53). Dimensions of healthy neutrophil nuclei were 10.3–29.3 × 1.7–4.5 (20.5 ± 3.9 × 2.7 ± 0.6) (n = 450), area of 25.1–58.1 (41.7 ± 7.4) µm2 (n = 450). Thus, infected neutrophils were slightly longer and wider, with larger surface area. Nuclei of infected neutrophils were slightly longer and narrower, with greater surface area.
Remarks
Prior to this study, Hepatozoon spp. reported from wild carnivores were only H. felis, H. canis and mostly as an unidentified species of Hepatozoon [6, 10,11,12, 22, 24, 25, 29, 49,50,51,52,53,54,55,56,57,58,59,60,61,62,63], except by Hodžić et al. [20] who recently described Hepatozoon silvestris Hodžić, Alić, Prašović, Otranto, Baneth & Duscher, 2017 in an European wildcat from eastern Europe. Infection with a morphologically and genetically distinct Hepatozoon sp. was confirmed by our study in wild African leopards in various areas throughout South Africa, both in male and female hosts. The haemogregarine described here appears to develop only gamont stages in the peripheral blood of P. p. pardus. Therefore, with no division stages detected, it was placed within the genus Hepatozoon.
Hepatozoon luiperdjie n. sp. was on average longer than H. canis from Cerdocyon thous (L.) in Brazil [51], and longer, but with a similar average width to H. felis from Felis catus (L.) in Israel [19], therefore within the relative morphometrical range of both species (Table 3). This haemogregarine was morphometrically most similar to an unnamed species of Hepatozoon detected in Lynx rufus (Schreber) from the USA [57]. It also differed from the other species of Hepatozoon detected in the monocytes of leopards during our study. The most striking feature of H. luiperdjie n. sp. is the densely chromatisized, acentric nucleus, relatively smaller than that of H. felis [19, 20]. Hepatozoon luiperdjie n. sp. also seemed to exclusively infect the neutrophils of the host. The life-cycle of this parasite remains to be determined.
Hepatozoon ingwe n. sp.
Type-host:Panthera pardus pardus (L.) (Carnivora: Felidae).
Type-locality: Lajuma Research Centre (23°02ʹ17.1ʺS, 29°26ʹ26.5ʺE), Limpopo Province, South Africa.
Other localities: Greater Kruger Conservation Area (24°34ʹ43.50ʺS, 31°25ʹ46.48ʺE), Mpumalanga Province; captive facility (24°30ʹ52.44ʺS, 30°54ʹ8.82ʺE), Mpumalanga Province; Lydenburg area (25°9ʹ51.31ʺS, 30°26ʹ55.46ʺE), Mpumalanga Province, South Africa.
Type-material: Hapantotype, 1 peripheral blood smear from the type-host P. p. pardus and type-locality (23°02ʹ17.1ʺS, 29°26ʹ26.5ʺE), deposited under accession number NMBP393 in the protozoan collection of the National Museum, Bloemfontein, South Africa.
Vector: Unknown.
Representative DNA sequences: Two sequences, representing a 995 nt fragment of the 18S rRNA gene of Hepatozoon ingwe n. sp., isolated from the type-host P. p. pardus, deposited under the accession numbers MN793000 and MN793001 in the GenBank database.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [48], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:9E65A924-729F-43A5-AE1F-8001204B6A6A.The LSID for the new name Hepatozoon ingwe n. sp. is urn:lsid:zoobank.org:act:65A2DC3D-BABB-443A-82A7-DBCFFCAE3636.
Etymology: The species epithet is derived from that of the Zulu language name for the host P. p. pardus, which in Zulu is referred to as “ingwe”. Noun in apposition.
Description
Gamonts. Most abundant stage in peripheral blood smears (Fig. 1g–k). Extracellular forms (Fig. 1i) and immature gamonts (Fig. 1j) rarely observed, no division stages detected. Mature gamonts measure 9.8–12.6 × 4.5–5.0 (11.4 ± 1.2 × 4.8 ± 0.2) (n = 87), surface area of 38.7–48.9 (44.2 ± 4.4) µm2 (n = 87); mostly visible within leukocyte cytoplasm (Fig. 1g, h, k), but in some cases gamonts were concealed by leukocyte nucleus; elongate with round extremities, cytoplasm stained pale blue, slight granulation, minimal basophilic stippling anteriorly; cytoplasm contained bright pink staining granules (Fig. 1g, h). Gamonts with thin visible capsules (Fig. 1g, thick arrow) and 2 to 4 prominent vacuoles posteriorly situated (Fig. 1h, thin arrow); gamont nuclei measure 4.4–5.7 × 2.5–3.7 (5.1 ± 0.6 × 3.0 ± 0.6) (n = 87), area of 8.7–16.5 (12.2 ± 3.3) µm2 (n = 87). Nuclei stained dark purple with loosely stranded chromatin, through which parts of cytoplasm were often visible (Fig. 1g, h). Nuclei elongate, usually narrower than gamont at widest point, mostly anteriorly located. Capsule measured 0.4–0.7 (0.5 ± 0.1) thick (n = 87) and observable in most gamonts.
Prevalence and parasitaemia. Detected in the peripheral blood of 7 out of 16 individual P. p. pardus sampled (prevalence of 44%). Of these 6 out of 16 leopards had co-infections with H. luiperdjie n. sp. (prevalence of 38%) and 1 out of 16 individuals was solely infected with this species of Hepatozoon (prevalence 6%). More males than females were infected, prevalence 50% (4/8) of males and 38% (3/8) of females. One captive female was infected (prevalence in captivity 13% or 1/8). Parasitaemia varied between individuals and could only be determined in WM1 (32%), WF1 (57%), WF3 (13%), WM4 (21%) and WM5 (7%). Average parasitaemia was 30.8%. Gamonts not observed in smears of CF5; however, prevalence confirmed by PCR amplification.
Effect on host cells. Gamonts usually compacted lymphocyte nuclei towards one side and completely usurped lymphocyte cytoplasm (Fig. 1g, h). Parasitized lymphocytes measured 10.9–11.6 × 8.4–9.7 (11.3 ± 0.3 × 9.1 ± 0.6) (n = 87), area of 73.6–83.4 (79.2 ± 4.3) µm2 (n = 87). Healthy, uninfected lymphocytes measured 6.8–15.7 × 6.1– 13.9 (10.9 ± 2.1 × 9.7 ± 1.9) (n = 261), area of 36.6–156.8 (81.7 ± 29.4) µm2 (n = 261). Nuclei of parasitized lymphocytes measured 8.4–11.9 × 3.8–5.7 (10.0 ± 1.5 × 4.78 ± 0.8) (n = 87), area of 30.2–36.9 (33.5 ± 3.0) µm2 (n = 87). Dimensions of healthy lymphocyte nuclei were 6.1–16.1 × 4.2–10.7 (9.7 ± 2.2 × 7.3 ± 1.4) (n = 261), area of 26.2–92.1 (56.2 ± 15.4) µm2 (n = 261). Thus, infected lymphocytes were slightly longer and narrower, with smaller surface area. Infected lymphocyte nuclei measured slightly longer and narrower, with smaller surface area.
Remarks
This haemogregarine appears to develop only gamont stages in the peripheral blood of P. p. pardus, and with no division stages detected it was placed within Hepatozoon. Hepatozoon ingwe n. sp. measured within the same range size as H. luiperdjie n. sp. described above. However, H. ingwe n. sp. seemed to exclusively infect the lymphocytes of the host, unlike the gamonts of H. luiperdjie n. sp. Hepatozoon ingwe n. sp. was morphometrically similar to an unnamed Hepatozoon detected in the neutrophils of P. p. ciscaucasica from Iran [12] (Table 3). This haemogregarine also measured longer and wider than H. felis from domestic cats in Israel [19], widely considered a redescription of H. felis (Table 3). Hepatozoon ingwe n. sp. was significantly longer (P = 0.0064) and somewhat wider (P = 0.3023), with a comparatively larger surface area than that of H. luiperdjie n. sp. (P = 0.0593) (Table 3). Characteristic features of H. ingwe n. sp. include the pale blue staining cytoplasm containing bright pink staining granules, prominent vacuoles at the posterior, and elongated nuclei similar to that of H. canis. The life-cycle of this parasite remains to be determined.
Differential diagnoses
Each species of haemogregarine described here infected a particular type of leukocyte: H. luiperdjie n. sp. were exclusively found in neutrophils and H. inwe n. sp. in lymphocytes. The parasitaemia of H. ingwe n. sp. (30.8%), and H. luiperdjie n. sp. (11.0%), was higher than the range of 0.1–4.0% reported by other studies on domestic cats in Israel [64], an ocelot Leopardus pardalis (L.) in Brazil [35], a Tsushima leopard cat Prionailurus bengalensis (Kerr) in Thailand [54], Iriomote cats P. b. iriomotensis (Imaizumi) in Japan [52] and a Tsushima leopard cat P. bengalensis in Japan. The parasitaemia of H. ingwe n. sp. was also higher than that of H. luiperdjie n. sp., possibly indicating that leopard host immune system may be better at suppressing infections by H. luiperdjie n. sp. in neutrophils, than H. ingwe n. sp. in lymphocytes. Additionally, there seemed to be a clear association between sex and parasitaemia in this study, with average parasitaemia in females (H. luiperdjie n. sp. parasitized 18.2% and H. ingwe n. sp. parasitized 35% of host cells) higher than that in males (H. luiperdjie n. sp. parasitized 5.99% and H. ingwe n. sp. parasitized 20% of host cells). This may be a noticeable health concern, since it is still unclear whether feline hepatozoonosis can be transferred within the uterus. However, this trend in parasitaemia needs further investigation over a larger variety and number of felid hosts.
In addition to infecting different types of host cells, the two new species showed clear morphological distinctions of peripheral blood gamont stages on a morphometric basis and differences in staining properties. The gamonts of H. luiperdjie n. sp. are shorter, thinner and with a smaller surface area than those of H. ingwe n. sp. Furthermore, the staining properties of their cytoplasm, the marked presence of vacuoles in H. ingwe n. sp. and the morphometric and staining differences in their nuclei. In terms of nucleus dimensions, the nucleus of H. luiperdjie n. sp. was significantly shorter (P < 0.0001), broader (P < 0.0001) and larger than that of H. ingwe n. sp. (P = 0.0025). Both species described here had a larger surface area compared to the unnamed species of Hepatozoon parasitizing neutrophils of Leopardus pardalis from Brazil [35] and an unnamed species of Hepatozoon parasitising neutrophils of P. p. ciscaucasica from Iran [12] (Table 3). While both new species of Hepatozoon had a smaller surface area as compared to an unnamed species of Hepatozoon from domestic cats [65] and H. canis from Cerdocyon thous from Brazil, respectively [61] (Table 3). Data on the morphometrics of large numbers of gamonts are scarce, with some studies measuring only a few gamonts [19, 20], or even only a single gamont [35]. Our study focused on screening live hosts, while most other studies analysed necropsied or biopsied samples [19, 20, 66], or did not focus on morphological descriptions of the gamont stage [10, 11, 22, 24, 27, 29, 31, 52, 53, 55, 58, 64, 67,68,69]. Several of these studies reported only on molecular detection due to the general low parasitaemia of these haemogregarines [66], as confirmed by the absence of gamonts in the peripheral blood smear of CF5 in our study.
Molecular analyses
Amplicons were derived from H. luiperdjie n. sp. and H. ingwe n. sp. from the blood of leopards and the details of all sequences used for analysis are presented in Table 2. Although eight sequences were obtained from infected leopards, only a single sequence per species of Hepatozoon was used in the phylogenetic analysis (Fig. 2), as sequences obtained from samples with single species infections were identical for the respective new species. Sequences obtained from leopards with mixed Hepatozoon infections were not included in the phylogenetic analysis, as sequences contained a double chromatogram peak or two separate bases called at the same position (heterozygous positions) from the two new species of Hepatozoon amplified. Based on the uncorrected p-distance for the 18S rRNA gene between H. ingwe n. sp. and H. felis [amplified from the Asiatic lion, Panthera leo persica (L.) (GenBank: HQ829440) and Indian leopard, Panthera pardus fusca (L.) (GenBank: HQ829444)] interspecific divergence was 1.0% (Additional file 2: Table S1). Hepatozoon luiperdjie n. sp. and H. felis amplified from domestic cats, F. catus (GenBank: AY620232 and AY628681) and from a Bengal tiger, P. tigris tigris (L.) (GenBank: HQ829445) had an interspecific divergence of 1.0% (Additional file 2: Table S1). The interspecific divergence between H. luiperdjie n. sp. and H. ingwe n. sp. was 1.0%.
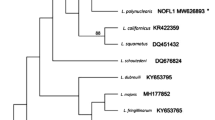
Bayesian inference (BI) phylogram based on 18S rDNA sequences. Phylogram illustrating the phylogenetic relationships between Hepatozoon luiperdjie n. sp. and Hepatozoon ingwe n. sp. (shown in bold) with 55 representative sequences of other species of Dactylosoma, Haemogregarina, Hepatozoon, Karyolysus and Hemolivia retrieved from GenBank. Adelina dimidiata, Adelina grylli and Klossiella equi were selected as the outgroup. Posterior probability values lower than 0.60 were omitted. The scale-bar represents 0.02 nucleotide substitutions per site. Distinct clades are presented in alternating colors and letters A to L highlight 12 distinct clades
For the BI phylogenetic analyses, first a large dataset was used comprising 297 sequences (Additional file 1: Figure S1). This analysis included all the available 18S rDNA sequence data of H. felis downloaded from GenBank. The genera Hemolivia, Hepatozoon (parasitising amphibian, reptilian and rodent hosts), Karyolysus, Haemogregarina and Dactylosoma formed separate and well-supported clades at the base of the phylogeny (Additional file 1: Figure S1). Species of Karyolysus and several species most likely incorrectly identified as species of Hepatozoon, formed a sister group to a large clade comprising species of Hepatozoon from large mammals. The phylogenetic analysis showed H. felis as paraphyletic forming several clusters, along with Hepatozoon americanum Vincent-Johnson, MacIntire, Lindsay, Lenz, Baneth, Shkap & Blagburn, 1997, Hepatozoon apri Yamamoto, Tokiwa, Tobiume, Akamatsu, Matsuo, Moribe & Ike, 2017, Hepatozoon canis, Hepatozoon martis Hodžić, Alić, Beck, Beck, Huber, Otranto, Baneth & Duscher, 2018, Hepatozoon silvestris, Hepatozoon ursi Kubo, Uni, Agatsuma, Nagataki, Panciera, Tsubota, Nakamura, Sakai, Masegi & Yanai, 2008 and H. luiperdjie n. sp. and H. ingwe n. sp. Although several sequences are identified or designated as H. felis, we consider the sequences isolated from H. felis in domestic cats from Spain (GenBank: AY620232; AY628681) as sufficient representatives of H. felis based on phylogenetic comparisons to sequences in the formal redescription and molecular characterisation of H. felis [19].
The second BI phylogenetic analysis was based on 57 18S rDNA sequences (Fig. 2). Species of Hepatozoon isolated from large mammal hosts formed a large well-supported clade (Clades A-E, Fig. 2). Clades A and B (monophyletic group), and Clades C, D and E, formed a polytomy of four distinct groups. In Clade A, H. ingwe n. sp. was shown as a sister taxon to a well-supported monophyletic cluster of H. felis in big cats from India, namely the Asiatic lion (GenBank: HQ829440) and Indian leopard (GenBank: HQ829444) (Clade A in Fig. 2). Furthermore, H. silvestris (GenBank: KX757032), isolated form the European wild cat Felis silvestris silvestris Schreber, and H. apri (GenBank: LC314791) isolated from the Japenese boar, Sus scrofa leucomystax Temminck formed a polytomy at the base of clade A. In clade B, H. luiperdjie n. sp. clustered with sequences isolated from H. felis in domestic cats from Spain (GenBank: AY620232; AY628681), a bengal tiger, P. tigris tigris (GenBank: HQ829445) from India, and H. felis amplified from a tick Rhipicephalus sanguineus (Latreille) that was collected from an Asiatic lion Panthera leo persica (GenBank: KY056823). Clade C was a monophyltic clade of H. canis and species of Hepatozoon isolated from various canid hosts. In clade D, H. martis isolated from the European badger Meles meles (L.) and the European pine marten Martes martes (L.), formed a sister group to H. ursi isolated from the Japanese black bear Ursus thibetanus japonicus Schlegel and the sloth bear Melursus ursinus (Shaw). Clade E comprised H. americanum, isolated from domestic dogs, Canis familiaris L. and an unnamed species of Hepatozoon in crab-eating foxes Dusicyon thous azarae (L.).
Discussion
Since the late 1960’s, the African leopard has been a favoured research subject of ethologists and ecologists. However, research on haematozoans of large carnivores is sparse and often occurs as non-specific reports, and according to Peirce et al. [6] researchers often automatically identify species of Hepatozoon in African carnivores as H. canis or H. felis. According to Baneth et al. [19], most studies on hepatozoonosis have emphasized the detection of the parasite, with little attention given to other aspects such as transmission and epidemiology. It is therefore not unexpected that the scant research on health aspects of African leopards left gaps in knowledge of especially their haemoparasites. Our study addressed these gaps and confirmed co-infection of two morphologically and genetically distinct Hepatozoon species in wild and captive African leopards in various areas throughout South Africa, both in male and female hosts.
No leopards sampled during our study displayed any clinical symptoms associated with hepatozoonosis, confirming similar reports by authors such as Brocklesby & Vidler [62], Averbeck et al. [22] and East et al. [25]. Prevalence of Hepatozoon varies between hosts and regions. The overall prevalence of hepatozoonosis recorded in this study was 56%, which is similar to that reported in Iriomote cats Prionailurus bengalensis iriomotensis from Japan (56.7%) [55], spotted hyenas Crocuta crocuta (Erxleben) from Zambia (56%) [28], and captive Asiatic lions P. l. persica from India (55.56%) [11]. Prevalence found during our study was slightly higher than the prevalence of unknown species of Hepatozoon reported from Indian leopards P. p. fusca (50%) [11] and much higher than that of African wild dogs Lycaon pictus (Temminck) in South Africa (0.7%) [31]. The prevalence of H. luiperdjie n. sp. (50%) was similar to that of an unknown species of Hepatozoon reported from Indian leopards P. p. fusca in India (50%) [11], and it was lower than the prevalence of H. ingwe n. sp. (44%). The prevalence of both new species described in this study was higher in males than females, with 63% of males infected with H. luiperdjie n. sp. and 50% of males infected with H. ingwe n. sp., and 38% of females infected with H. luiperdjie n. sp. and H. ingwe n. sp., respectively.
The two new haemogregarines had dissimilar effects on their respective host cells. Hepatozoon luiperdjie n. sp. caused enlargement of neutrophil cells and their nuclei and H. ingwe n. sp. reduced the size of lymphocytes and condensed their nuclei. Although Baneth et al. [19] reported on a species of Hepatozoon that infects both neutrophils and lymphocytes, our study showed that co-infecting species of Hepatozoon can inhabit different types of leukocytes, with different effects and morphological characteristics of their gamont stages.
It is evident based on morphological and molecular data that H. luiperdjie n. sp. and H. ingwe n. sp. are distinct species. These species are also distantly related to H. felis (GenBank: AY628681) based on 18S rDNA sequence comparisons, isolated by Criado-Fornelio et al. [50] from domestic cats from Spain. The H. felis isolates from Spain [50] are widely regarded as the representative H. felis isolates to be used for comparison [25, 55, 60]. Thus, based on the phylogenetic relationships and comparisons of H. felis and H. felis-like species of Hepatozoon, the identity of the Hepatozoon infecting large carnivores, currently identified as H. felis by Pawar et al. [11] is questioned. Therefore, based on these genetic distinctions, as well as morphological characteristics and effect on host cells, the two haemogregarines described here were deemed to be two different species and new to science. The phylogenetic results from our study showed H. luiperdjie n. sp. and H. ingwe n. sp. as distinct species as compared to the currently recognised species of Hepatozoon infecting large mammal hosts, i.e. H. americanum, H. apri, H. canis, H. felis, H. martis, H. silvestris and H. ursi. We therefore suggest that these haemogregarines may need to be re-classified based on morphological, morphometric and molecular analysis.
Prior to our study, only H. felis, H. canis and several unknown species of Hepatozoon have been reported from African carnivores [6, 8, 10, 22, 24, 28, 31, 57, 64, 65], but this study confirmed a mixed population of two genetically distinct haemogregarines from both captive and wild leopards, across males and females and from leopards representative from three core populations as identified by Daly et al. [36]. The topology of our BI tree confirmed the suggestion of Hodžić et al. [20], that H. felis should be viewed as a species complex.
Conclusions
As shown in this study, morphology and the effect on host cells are important parameters that should be taken into account when identifying species of Hepatozoon. By using different techniques of identification, a better understanding of the parasite and its relation to its host may become possible. Thus, Hepatozoon species identified as either H. felis or H. canis based on the host parasitized should be re-evaluated using both morphological and molecular characteristics, as well as the type of and effect on the host cells. The value of the results from our study, in addition to describing two new haemogregarine species, is that we present results obtained from live animals in the wild. It is important to identify these parasites to species level in order to better understand potential zoonotic effects on different host species, and to further investigate the possible transfer of haemogregarines from wild to domestic animals. In addition, this paper provides valuable criteria to be considered when describing Hepatozoon infections from wild carnivores. Possible future work on these haemogregarines should include in depth investigations on the life cycles and vectors of these species.
Availability of data and materials
Data supporting the conclusions of this article are included within the article and its additional files. The datasets used and analysed during this study will be made available by the corresponding author upon reasonable request. The newly generated sequences were submitted to the GenBank database under the accession numbers MN792996-MN792999 (Hepatozoon spp. mixed infections), MN792000-MN792001 (H. ingwe n. sp.), and MN792002-MN792004 (H. luiperdjie n. sp.).
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- BI:
-
Bayesian inference
- BIC:
-
Bayesian information criterion
- GTR+I+G:
-
general time reversible model with estimates of invariable sites and a discrete Gamma distribution
- ICZN:
-
International Code of Zoological Nomenclature
- IUCN:
-
International Union for the Conservation of Nature
- LSID:
-
Life Science Identifier
- MCMC:
-
Markov Chain Monte Carlo
- NCBI:
-
National Center for Biotechnology Information
- PAUP:
-
Phylogenetic Analysis Using Parsimony
- PCR:
-
polymerase chain reaction
- NRF:
-
South African National Research Foundation
- UESM:
-
Unit for Environmental Sciences and Management
References
Smith TG. The genus Hepatozoon (Apicomplexa: Adeleorina). J Parasitol. 1996;82:565–85.
Patton WS. The haemogregarines of mammals and reptiles. Parasitology. 1908;1:318–21.
Wenyon CM. Protozoology: a manual for medical men, veterinarians and zoologists. London: Baillere, Tindall and Cox Ltd.; 1926.
Beaufils JP, Martin-Granel J, Jumelle P. Hepatozoon spp. parasitemia and feline leukemia virus infection in two cats. Feline Pract. 1998;26:10–3.
Levine ND. The protozoan phylum Apicomplexa. Boca Raton, USA: CRC Press; 1988.
Peirce MA, Laurenson MK, Gascoyne SC. Hepatozoonosis in cheetahs and wild dogs in the Serengeti ecosystem. Afr J Ecol. 1995;33:273–5.
Murray DL, Kapke CA, Evermann JF, Fuller TK. Infectious disease and the conservation of free-ranging large carnivores. Anim Conserv. 1999;2:241–54.
Brocklesby DW, Vidler BO. Some new host records for Hepatozoon species in Kenya. Vet Record. 1963;75:1265.
Keymer IF. Blood protozoa of wild carnivores in Central Africa. J Zool. 1971;164:513–24.
McCully RM, Basson PA, Bigalke RD, DeVoss V, Young E. Observations on naturally acquired hepatozoonosis of wild carnivores and dogs in the Republic of South Africa. Onderstepoort J Vet Res. 1975;42:117–34.
Pawar RM, Poornachandar A, Srinivas P, Rao KR, Lakshmikantan U, Shivaji S. Molecular characterization of Hepatozoon spp. infection in endangered Indian wild felids and canids. Vet Parasitol. 2012;186:475–9.
Khoshnegah J, Mohri M, Mirshahi A, Mousavi SJ. Detection of Hepatozoon sp. in a Persian leopard (Panthera pardus ciscaucasica). J Wildl Dis. 2012;48:776–80.
Davies AJ, Johnston MRL. The biology of some intraerythrocytic parasites of fishes, amphibians and reptiles. Adv Parasitol. 2000;45:1–107.
Perkins SL, Keller AK. Phylogeny of nuclear small subunit rRNA genes of hemogregarines amplified with specific primers. J Parasitol. 2001;87:870–6.
Netherlands EC, Cook CA, Kruger DJ, du Preez LH, Smit NJ. Biodiversity of frog haemoparasites from sub-tropical northern Kwazulu-Natal, South Africa. Int J Parasitol Parasites Wildl. 2015;4:135–41.
Netherlands EC, Cook CA, Du Preez LH, Vanhove MPM, Brendonck L, Smit NJ. Monophyly of the species of Hepatozoon (Adeleorina: Hepatozoidae) parasitizing (African) anurans, with the description of three new species from hyperoliid frogs in South Africa. Parasitology. 2017;145:1039–50.
Van As J, Davies AJ, Smit NJ. Hepatozoon langii n. sp. and Hepatozoon vacuolatus n. sp. (Apicomplexa: Adeleorina: Hepatozoidae) from the crag lizard (Sauria: Cordylidae) Pseudocordylus langi from the North Eastern Drakensberg escarpment, Eastern Free State, South Africa. Zootaxa. 2013;3608:345–56.
Borges-Nojosa DM, Borges-Leite MJ, Maia JP, Zanchi-Silva D, da Rocha Braga R, Harris DJ. A new species of Hepatozoon Miller, 1908 (Apicomplexa: Adelerina) from the snake Philodryas nattereri Steindachner (Squamata: Dipsadidae) in northeastern Brazil. Syst Parasitol. 2017;94:65–72.
Baneth G, Sheiner A, Eyal O, Hahn S, Beaufils J-P, Anug Y, et al. Redescription of Hepatozoon felis (Apicomplexa: Hepatozoidae) based on phylogenetic analysis, tissue and blood form morphology, and possible transplacental transmission. Parasit Vectors. 2013;6:1–10.
Hodžić A, Alić A, Prašović S, Otranto D, Baneth G, Duscher GG. Hepatozoon silvestris sp. nov.: morphological and molecular characterization of a new species of Hepatozoon (Adeleorina: Hepatozoidae) from the European wild cat (Felis silvestris silvestris). Parasitology. 2017;144:650–61.
Brocklesby DW. Illustrations of a Hepatozoon species in the heart of a lion. J Zool. 1971;164:525–8.
Averbeck A, Bjork KE, Packer C, Herbst L. Prevalence of hematozoans in lions (Panthera leo) and cheetah (Acinonyx jubatus) in Serengeti National Park and Ngorongoro Crater, Tanzania. J Wildl Dis. 1990;26:392–4.
Dubey JP, Bwangamoi O. Microbesnoitia leoni Bwangamoi, 1989, from the African lion (Panthera leo) redetermined as a junior synonym of Hepatozoon canis (James, 1905) Wenyon, 1926. J Parasitol. 1994;80:333–4.
Van Heerden J, Mills MG, Van Vuuren MJ, Kelly PJ, Dreyer MJ. An investigation into the health status and diseases of wild dogs (Lycaon pictus) in the Kruger National Park. J S Afr Vet Assoc. 1995;66:18–27.
East ML, Wibbelt G, Lieckfeldt D, Ludwig A, Goller K, Wilhelm K, et al. A Hepatozoon species genetically distinct from H. canis infecting spotted hyenas in the Serengeti ecosystem, Tanzania. J Wildl Dis. 2008;44:45–52.
Jittapalapong S, Rungphisutthipongse O, Maruyama S, Schaefer JJ, Stich RW. Detection of Hepatozoon canis in stray dogs and cats in Bangkok, Thailand. Ann N Y Acad Sci. 2006;1081:479–88.
Rubini AS, Paduan KS, Perez RR, Ribolla PEM, O’Dwyer LH. Molecular characterization of feline Hepatozoon species from Brazil. Vet Parasitol. 2006;137:168–71.
Williams BM, Berentsen A, Shock BC, Teixiera M, Dunbar MR, Becker MS, et al. Prevalence and diversity of Babesia, Hepatozoon, Ehrlichia, and Bartonella in wild and domestic carnivores from Zambia, Africa. Parasitol Res. 2014;113:911–8.
André MR, Adania CH, Teixeira RHF, Vargas GH, Falcade M, Sousa L, et al. Molecular detection of Hepatozoon spp. in Brazilian and exotic wild carnivores. Vet Parasitol. 2010;173:134–8.
Farkas R, Solymosi N, Takács N, Hornyák Á, Hornok S, Nachum-Biala Y, et al. First molecular evidence of Hepatozoon canis infection in red foxes and golden jackals from Hungary. Parasit Vectors. 2014;7:303.
Matjila PT, Leisewitz AL, Jongejan F, Bertschinger HJ, Penzhorn BL. Molecular detection of Babesia rossi and Hepatozoon sp. in African wild dogs (Lycaon pictus) in South Africa. Vet Parasitol. 2008;157:123–7.
Penzhorn BL, Cook CA, Smit NJ, Vorster I, Harrison-White RF, Oosthuizen MC. Occurrence of Hepatozoon canis (Adeleorina: Hepatozoidae) and Anaplasma spp. (Rickettsiales: Anaplasmataceae) in black-backed jackals (Canis mesomelas) in South Africa. Parasit Vectors. 2018;11:158.
Barta JR, Ogedengbe JD, Martin DS, Smith TG. Phylogenetic position of the adeleorinid Coccidia (Myzozoa, Apicomplexa, Coccidia, Eucoccidiorida, Adeleorina) inferred using 18S rDNA sequences. J Eukaryot Microbiol. 2012;59:171–80.
Cook CA, Netherlands EC, Smit NJ. First Hemolivia from southern Africa: reassigning chelonian Haemogregarina parvula Dias, 1953 (Adeleorina: Haemogregarinidae) to Hemolivia (Adeleorina: Karyolysidae). Afr Zool. 2015;50:165–73.
Metzger B, dos Santos Paduan K, Rubini AS, de Oliveira TG, Pereira C, O’Dwyer LH. The first report of Hepatozoon sp. (Apicomplexa: Hepatozoidae) in neotropical felids from Brazil. Vet Parasitol. 2008;152:28–33.
Daly B, Power J, Camacho G, Traylor-Holzer K, Barber S, Catterall S, et al. Leopard (Panthera pardus) population and habitat viability assessment. In: Proceedings of a workshop of the conservation breeding specialist group (World Conservation Union (IUCN) Species Survival Commission). Endangered Wildlife Trust, Johannesburg. IUCN, Johannesburg, South Africa; 2005.
Fattebert J, Balme G, Dickerson T, Slotow R, Hunter L. Density-dependent natal dispersal patterns in a leopard population recovering from over-harvest. PLoS ONE. 2015;10:e0122355.
Stander PE. Field age determination of leopards by tooth wear. Afr J Ecol. 1997;35:156–61.
Balme G, Hunter L, Britz NDW. A case of offspring adoption in leopards, Panthera pardus. Afr J Wildl Res. 2012;42:63–6.
Rasband WS. 2014. ImageJ. U. S. National Institutes of Health, Bethesda, Maryland, USA. 2014; http://imagej.nih.gov/ij.
Mathew JS, Van Den Bussche RA, Ewing SA, Malayer JR, Latha BR, Panciera RJ. Phylogenetic relationships of Hepatozoon (Apicomplexa: Adeleorina) based on molecular, morphologic, and life-cycle characters. J Parasitol. 2000;86:366–72.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Darriba D, Taboada GL, Doallo R, Posada D. Jmodeltest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures Math Life Sci. 1986;17:57–86.
Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5.
ICZN. International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull Zool Nomencl. 2012;69:161–9.
Kubo M, Miyoshi N, Yasuda N. Hepatozoonosis in two species of Japanese wild cat. J Vet Med Sci. 2006;68:833–7.
Criado-Fornelio A, Ruas JL, Casado N, Farias NA, Soares MP, Müller G, et al. New molecular data on mammalian Hepatozoon species (Apicomplexa: Adeleorina) from Brazil and Spain. J Parasitol. 2006;92:93–9.
Waner T, Baneth G, Zuckerman A, Nyska A. Hepatozoon canis: size measurement of gametocyte using image analysis technology. Comp Haematol Int. 1994;4:177–9.
Tateno M, Nishio T, Matsuo T, Sakuma M, Nakanishi N, Izawa M, et al. Epidemiological survey of tick-borne protozoal infection in Iriomote cats and Tsushima leopard cats in Japan. J Vet Med Sci. 2013;75:985–9.
Salakij C, Prihirunkit K, Narkkong NA, Apibal S, Tongthainun D. Hematology, cytochemistry and ultrastructure of blood cells in clouded leopard (Neofelis nebulosa). J Anim Vet Adv. 2008;7:847–53.
Salakij C, Sirinarumitr T, Tongthainun D. Molecular characterization of Hepatozoon species in a leopard cat (Prionailurus bengalensis) from Thailand. Vet Clin Pathol. 2010;39:199–202.
Sakuma M, Nishio T, Nakanishi N, Izawa M, Asari Y, Okamura M, et al. A Case of Iriomote cat (Prionailurus bengalensis iriomotensis) with Hepatozoon felis parasitemia. J Vet Med Sci. 2011;73:1381–4.
Mercer SH, Jones LP, Rappole JH, Twedt D, Laack LL, Craig TM. Hepatozoon sp. in wild carnivores in Texas. J Wildl Dis. 1988;24:574–6.
Lane JR, Kocan AA. Hepatozoon infection in bobcats. J Am Vet Med Assoc. 1983;183:1323–4.
Kubo M, Jeong A, Kim SI, Kim YJ, Lee H, Kimura J, Agatsuma T, Sakai H, Yanai T. The first report of Hepatozoon species infection in leopard cats (Prionailurus bengalensis) in Korea. J Parasitol. 2010;96:437–9.
Kelly P, Marabini L, Dutlow K, Zhang J, Loftis A, Wang C. Molecular detection of tick-borne pathogens in captive wild felids, Zimbabwe. Parasit Vectors. 2014;7:514.
Davis S, Robinson RM, Craig TM. Naturally occurring hepatozoonosis in a coyote. J Wildl Dis. 1978;14:244–6.
Alencar NX, Kohayagawa A, Santarem VA. Hepatozoon canis infection of wild carnivores in Brazil. Vet Parasitol. 1997;70:279–82.
Brocklesby DW, Vidler BO. Some parasites of East African wild animals. E Afr Wildl J. 1965;3:120–2.
Okubanjo OO, Adeshina OA, Jatau ID, Natala AJ. Prevalence of Babesia canis and Hepatozoon canis in Zaria, Nigeria. Sokoto J Vet Sci. 2013;11:15–20.
Baneth G, Aroch I, Tal N, Harrus S. Hepatozoon species infection in domestic cats: a retrospective study. Vet Parasitol. 1998;79:123–33.
Perez RR, Rubini AS, O’Dwyer LH. The first report of Hepatozoon spp (Apicomplexa, Hepatozoidae) in domestic cats from São Paulo State, Brazil. Parasitol Res. 2004;94:83–5.
Harris DJ, Halajian A, Santos JL, Swanepoel LH, Taylor PJ, Xavier R. Diversity of haemoprotozoan parasites infecting the wildlife of South Africa. Folia Parasit. 2017;65:1–8.
Maddock A, Anderson A, Carlisle F, Galli N, James A, Verster A, Whitfield W. Changes in lion numbers in Hluhluwe-Umfolozi Park. Lammergeyer. 1996;44:6–18.
Ortuño A, Castellà J, Criado-Fornelio A, Buling A, Barba-Carretero JC. Molecular detection of a Hepatozoon species in stray cats from a feline colony in north-eastern Spain. Vet J. 2007;177:134–5.
Criado-Fornelio A, Buling A, Casado N, Gimenez C, Ruas J, Wendt L, et al. Molecular characterization of arthropod-borne hematozoans in wild mammals from Brazil, Venezuela and Spain. Acta Parasitol. 2009;54:187–93.
Kopecná J, Jirků M, Oborník M, Tokarev YS, Lukes J, Modrý D. Phylogenetic analysis of coccidian parasites from invertebrates: search for missing links. Protist. 2016;157:173–83.
Criado-Fornelio A, Rey-Valeiron C, Buling A, Barba-Carretero JC, Jefferies R, Irwin P. New advances in molecular epizootiology of canine hematic protozoa from Venezuela, Thailand and Spain. Vet Parasitol. 2007;144:261–9.
Sloboda M, Kamler M, Bulantová J, Votýpka J, Modrý D. A new species of Hepatozoon (Apicomplexa: Adeleorina) from Python regius (Serpentes: Pythonidae) and its experimental transmission by a mosquito vector. J Parasitol. 2007;93:1189–98.
Kubo M, Uni S, Agatsuma T, Nagataki M, Panciera RJ, Tsubota T, Nakamura S, Sakai H, Masegi T, Yanai T. Hepatozoon ursi n. sp. (Apicomplexa: Hepatozoidae) in Japanese black bear (Ursus thibetanus japonicus). Parasitol Int. 2008;57:287–94.
Maia JP, Harris DJ, Perera A. Molecular survey of Hepatozoon species in lizards from North Africa. J Parasitol. 2011;97:513–7.
Pawar RM, Poornachandar A, Shanmugam Arun A, Manikandan S, Shivaji S. Molecular prevalence and characterization of Hepatozoon ursi infection in Indian sloth bears (Melursus ursinus). Vet Parasitol. 2011;186:475–9.
de Bortoli CP, André MR, Braga MSC, Machado RZ. Molecular characterization of Hepatozoon sp. in cats from São Luís Island, Maranhão, northeastern Brazil. Parasitol Res. 2011;109:1189–92.
Tomé B, Maia JP, Harris DJ. Molecular assessment of apicomplexan parasites in the snake Psammophis from North Africa: do multiple parasite lineages reflect the final vertebrate host diet? J Parasitol. 2013;99:883–7.
Dvořáková N, Kvičerová J, Papoušek I, Javanbakht H, Tiar G, Široký P. Haemogregarines from Western Palaearctic freshwater turtles (genera Emys, Mauremys) are conspecific with Haemogregarina stepanowi Danilewsky, 1885. Parasitology. 2013;141:522–30.
Kvičerová J, Hypša V, Dvořáková N, Mikulíček P, Jandzik D, Gardner MG, et al. Hemolivia and Hepatozoon: haemogregarines with tangled evolutionary relationships. Protist. 2014;165:688–700.
Haklová-Kočíková B, Hižňanová A, Majláth I, Račka K, Harris DJ, Földvári G, et al. Morphological and molecular characterization of Karyolysus—a neglected but common parasite infecting some European lizards. Parasit Vectors. 2014;7:555.
Maia JP, Crottini A, Harris DJ. Microscopic and molecular characterization of Hepatozoon domerguei (Apicomplexa) and Foleyella furcata (Nematoda) in wild endemic reptiles from Madagascar. Parasite. 2014;21:47.
Dvořáková N, Kvičerová J, Hostovský M, Široký P. Haemogregarines of freshwater turtles from Southeast Asia with a description of Haemogregarina sacaliae sp. n. and a redescription of Haemogregarina pellegrini Laveran and Pettit, 1910. Parasitology. 2015;142:816–26.
Karadjian G, Chavatte JM, Landau I. Systematic revision of the adeleid haemogregarines, with creation of Bartazoon n. g., reassignment of Hepatozoon argantis Garnham, 1954 to Hemolivia, and molecular data on Hemolivia stellata. Parasite. 2015;22:31.
Barandika JF, Espí A, Oporto B, Del Cero A, Barral M, Povedano I, et al. Occurrence and genetic diversity of piroplasms and other apicomplexa in wild carnivores. Parasitology. 2016;2:1–7.
Cook CA, Netherlands EC, Smit NJ. Redescription, molecular characterisation and taxonomic re-evaluation of a unique African monitor lizard haemogregarine Karyolysus paradoxa (Dias, 1954) n. comb. (Karyolysidae). Parasit Vectors. 2016;9:347.
Bhusri B, Sariya L, Mongkolphan C, Suksai P, Kaewchot S, Changbunjong T. Molecular characterization of Hepatozoon felis in Rhipicephalus sanguineus ticks infested on captive lions (Panthera leo). J Parasit Dis. 2017;41:903–7.
Yamamoto M, Tokiwa T, Tobiume M, Akamatsu S, Matsuo K, Moribe J, et al. Hepatozoon apri n. sp. (Adeleorina: Hepatozoidae) from the Japanese wild boar Sus scrofa leucomystax (Mammalia: Cetartiodactyla). Int J Parasitol Parasites Wildl. 2017;6:354–60.
Hodžić A, Alić A, Beck R, Beck A, Huber D, Otranto D, et al. Hepatozoon martis n. sp. (Adeleorina: Hepatozoidae): morphological and pathological features of a Hepatozoon species infecting martens (family Mustelidae). Ticks Tick Borne Dis. 2018;9:912–20.
Cook CA, Netherlands EC, Van As J, Smit NJ. Two new species of Hepatozoon (Apicomplexa: Hepatozoidae) parasitising species of Philothamnus (Ophidia: Colubridae) from South Africa. Folia Parasitol. 2018;65:004.
Léveillé AN, Bland SK, Carlton K, Larouche CB, Kenney DG, Brouwer ER, et al. Klossiella equi infecting kidneys of Ontario horses: life cycle features and multi locus sequence based genotyping confirm the genus Klossiella belongs in the Adeleorina (Apicomplexa: Coccidia). J Parasitol. 2019;105:29–40.
Léveillé AN, Baneth G, Barta JR. Next generation sequencing from Hepatozoon canis (Apicomplexa: Coccidia: Adeleorina): complete apicoplast genome and multiple mitochondrion-associated sequences. Int J Parasitol. 2019;49:375–87.
Eljadar MSM, Singla LD, Mustafa RAA, Uppal SK. Morphometric variations in gametocytes of Hepatozoon canis from naturally infected dogs. J Parasit Dis. 2013;37:143–7.
Acknowledgements
We would like to thank all the facilities that allowed us access to sites for sample collection. In addition, we want to thank the Unit for Environmental Sciences and Management (UESM), (NWU-PC), for the use of their facilities and for funding for this project.
Funding
The financial assistance of the South African National Research Foundation (NRF) towards this research is hereby acknowledged (NRF project IFR1702100222411, Grant 109352 to NJS, PI). Opinions expressed and conclusions arrived at, are those of the authors and are not necessarily to be attributed to the NRF.
Author information
Authors and Affiliations
Contributions
MVA and NJS conceived and designed the study. MVA collected the blood samples and analysed morphometric data. MVA and ECN performed molecular work. MVA, ECN and NJS analysed molecular data. MVA wrote the manuscript, with input from ECN and NJS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study received ethical approval (North-West University ethics approval no. NWU-00255-17-S5).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Bayesian inference (BI) phylogram based on 297 18S rDNA sequences illustrating the phylogenetic relationships between H. felis, H. luiperdjie n. sp. and H. ingwe n. sp. (shown in bold) and other species of Dactylosoma, Haemogregarina, Hepatozoon, Karyolysus and Hemolivia retrieved from GenBank. Adelina dimidiate, A. grylli and Klossiella equi were selected as the outgroup. The scale-bar represents 0.02 nucleotide substitutions per site.
Additional file 2: Table S1.
Estimates of divergence using partial 18S rDNA sequences from the haemogregarines species used in the current study. Distance matrix showing ranges as percentage for the genetic p-distances between the sequences. Alignment length 1924 nt.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
van As, M., Netherlands, E.C. & Smit, N.J. Molecular characterisation and morphological description of two new species of Hepatozoon Miller, 1908 (Apicomplexa: Adeleorina: Hepatozoidae) infecting leukocytes of African leopards Panthera pardus pardus (L.). Parasites Vectors 13, 222 (2020). https://doi.org/10.1186/s13071-020-3933-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-3933-6