Abstract
Background
Rickettsia spp. are human pathogens that cause a number of diseases and are transmitted by arthropods, such as ixodid ticks. Estonia is one of few regions where the distribution area of two medically important tick species, Ixodes persulcatus and I. ricinus, overlaps. The nidicolous rodent-associated Ixodes trianguliceps has also recently been shown to be present in Estonia. Although no data are available on human disease(s) caused by tick-borne Rickettsia spp. in Estonia, the presence of three Rickettsia species in non-nidicolous ticks has been previously reported. The aim of this study was to detect, identify and partially characterize Rickettsia species in nidicolous and non-nidicolous ticks attached to rodents in Estonia.
Results
Larvae and nymphs of I. ricinus (n = 1004), I. persulcatus (n = 75) and I. trianguliceps (n = 117), all removed from rodents and shrews caught in different parts of Estonia, were studied for the presence of Rickettsia spp. by nested PCR. Ticks were collected from 314 small animals of five species [Myodes glareolus (bank voles), Apodemus flavicollis (yellow necked mice), A. agrarius (striped field mice), Microtus subterranius (pine voles) and Sorex araneus (common shrews)]. Rickettsial DNA was detected in 8.7% (103/1186) of the studied ticks. In addition to identifying R. helvetica, which had been previously found in questing ticks, we report here the first time that the recently described I. trianguliceps-associated Candidatus Rickettsia uralica has been identified west of the Ural Mountains.
Graphical Abstract

Similar content being viewed by others
Background
Rickettsia is a genus of small, obligate intracellular Gram-negative bacteria. Based on genomic analyses they are classified into four groups: the spotted fever group (SFG), the typhus group, the ancestral group and the transitional group [1]. Some SFG rickettsiae are transmitted by ticks of the family Ixodidae [2], and transmission may occur transovarially as well as transstadially [3, 4]. Several agents of tick-borne rickettsioses are known to circulate in Europe, including Rickettsia conorii, R. massiliae, R. slovaca, R. raoultii, R. monacensis and R. helvetica [2, 5], of which the last-mentioned is a frequently detected species in numerous Ixodidae ticks, including Ixodes ricinus, I. persulcatus, I. trianguliceps and Dermacentor reticulatus [2, 6, 7]. Although R. helvetica is not believed to be highly pathogenic to humans, several reports from Sweden [8, 9], the Netherlands [10], France and Italy [11] describe rash, mild fever, febrile illness, meningitis and other clinical symptoms associated with this agent in patients. In Estonia, Katargina et al. [12] reported the wide distribution of R. helvetica, as well as the presence of R. monacensis and Candidatus Rickettsia (Ca. R.) tarasevichiae, in questing ticks, but no human cases due to R. helvetica infection nor to the other two species had been reported at that time (2015).
Research on the circulation of Rickettsia spp. is still ongoing, btoh in vectors, which are mainly fleas and ticks, and in the latter’s main hosts (small mammals, wild and domestic animals). This is fairly wide research area, and new species are constantly being discovered, such as I. trianguliceps-associated Ca. R. uralica found in the Ural Mountains in Russia [7]. Also, the wide distribution of some types of vectors increases the probability of the prevalence of vector-associated pathogens, such as Ca. R. tarasevichiae, that have been found in China and Europe [12,13,14].
Current methods for disease surveillance include, among others, studying sentinel populations for the presence of pathogens in nature. Dogs [15] and cats [16] can be used as sentinels for rickettsiae. However, vectors can also serve as epidemiological sentinels [17].
Recent studies show that Ixodes spp. ticks can serve not only as vectors but also as the reservoir host of R. helvetica. At the present time there is no clear understanding of whether an mammal species is the host of R. helvetica, but rickettsial DNA has been found in the blood of wild animals, such as rodents, roe deer and wild boar [18], and domestic animals, such as dogs and cats [19]. Moreover, Burri et al. [20] reported negative results on R. helvetica xenodiagnostic as well as a low percentage of Rickettsia spp. from the positive host described by Tommassone et al. [21]. It can only be assumed that mammals can be potential hosts and that they may affect the natural transmission and distribution of rickettsiae.
The aim of the present study was to investigate the presence of Rickettsia spp. in ticks collected from small mammals.
Methods
Sample collection, species identification and DNA extraction
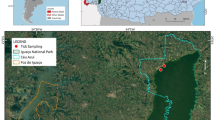
The study was performed retrospectively on 1186 ticks that had been removed from small mammals. The samples were collected at five sampling sites in Estonia, located in four mainland counties, namely Järvamaa, Lääne-Virumaa, Tartumaa (collecting in 2013 and 2014) and Pärnumaa (collecting was performed only in 2012), and in one island county, Saaremaa (Fig. 1). Live-trappings of mice, voles and shrews were carried out once a month during April–November 2012–2014 in natural habitats using Sherman LFA perforated live-traps (Ethical Committee Permission No. 124 by Estonian Ministry of Agriculture). Ten permanent stations (5 traps each station within a 2-m radius) were placed 100 m apart along a linear transect that randomly intersected different habitats (forest and semi-open). Trapping was performed during the nighttime, with the traps were set at 8 p.m. checked for animal the following morning at around 8 a.m. Bread was used as main bait method and vegetables served as water replenishment. The trapped animals were first identified to the species level and then killed by cervical dislocation by a specially trained person in accordance with Federation of European Laboratory Animal Science Association (FELASA) guidelines. Each animal was individually examined for the presence of ectoparasites, which were then removed, fixed in ethanol and stored at − 20 °C in separate tubes until further use. Any endangered species caught were immediately released in the habitat. For safety purposes, protective gloves and face masks were worn at all times while handling wild animals.
Trapping sites of small mammals during 2012–2014 in Estonia. Color of trapping sites indicated the study years: blue, trapping sites in 2013 and 2014; red, trapping sites in 2012; purple, trapping site in 2012–2014. The coordinates of the trapping sites are: Järvamaa (58.7365°N; 25.6682°E), Lääne-Virumaa (59.2260°N; 26.1335°E), Tartumaa (58.2493°N; 27.3023°E), Pärnumaa (58.0687°N; 24.8433°E) and Saaremaa (58.5075°N; 22.4107°E)
DNA was extracted from ticks using an ammonium hydroxide solution according to Moran-Cadenas et al. [22]. Tick species were identified by an internal transcribed spacer 2 gene (ITS2)-based multiplex PCR assay, as previously described by Värv et al. [23]. Only ticks identified at the species level based on ITS2 multiplex PCR results were included in this study, and ticks whose species identity remained undetermined were not used in the study.
Rickettsia spp. screening and genospecies detection
All ticks identified at the species level were screened individually by a nested PCR targeting a 667-bp fragment of the Rickettsia spp. citrate synthase A gene (gltA) using primers glt1–4, as described by Igolkina et al. [7], with subsequent sequencing of all positive samples. For samples identified as Ca. R. uralica and randomly selected samples identified as R. helvetica by initial screening, additional PCR amplification of ~ 770-bp fragment of the outer membrane protein B gene (ompB) was performed with primers 120-2788F and 120-3599R under conditions described previously [24]. Additionally, a subset of the latter samples was amplified by nested PCR of an 834-bp fragment of the cell-surface antigen 4 gene (sca4) with primers sc4-1 and Rj2837r for the primary reaction, and sc4-3 and sc4-4 for the nested reaction, as described by Igolkina et al. [7]. PCR products of all positive samples were sent for direct sequencing to the core laboratory of the Estonian Biocentre (Tartu, Estonia), followed by nucleotide sequence alignment using BioEdit v7.2.5 (Ibis Biosciences, Carlsbad, CA, USA) and genospecies identification with BLASTN® tools (http://www.ncbi.nlm.nih.gov/BLAST.cgi).
Results
Rickettsia screening and Rickettsia species
In this study 1186 ticks were collected from 314 small animals belonging to five mammalian species: Myodes glareolus, Apodemus flavicollis, A. agrarius, Microtus subterranius and Sorex araneus (Table 1; Additional file 1: Table S1). A total of 993 I. ricinus (924 larvae and 69 nymphs; from all 5 mammalian species), 117 I. trianguliceps (93 larvae and 24 nymphs, from My. glareolus and A. flavicollis) and 76 I. persulcatus (64 larvae and 12 nymphs, from My. glareolus, A. flavicollis and M. subterranius) were studied for presence of Rickettsia spp. (Table 2).
Rickettsial DNA was detected in 8.7% (103/1186) of the studied ticks, with positivity rates between tick species varying from zero for I. persulcatus to 3.4% (4/117) for I. trianguliceps to 10.0% (99/993) for I. ricinus (Table 2). As animal samples were not analyzed for the presence of Rickettsia spp., it is unknown whether ticks acquired the pathogen via transstadial or transovarial transmission, co-feeding or blood meal.
Rickettsial DNA was detected in ticks from all study sites, with the lowest positivity rates in Tartumaa and Saaremaa counties (2.6 and 4.8%, respectively) and the highest rate of 19.4% in Pärnumaa county.
Rickettsia spp. DNA was detected in ticks collected from 56 of 314 animals belonging to three species, namely My. glareolus (21.8%; 36/165), A. flavicollis (13.5%; 19/141) and S. araneus (33.3%; 1/3) (Additional file 1: Table S1). The number of ticks analyzed from a single animal varied from 1 to 32, while the rates of Rickettsia-positive ticks varied from 4.8 to 100%. The highest positivity rate of rickettsial DNA was observed in ticks from My. glareolus caught in Pärnumaa county (23.8%) (Table 1).
Partial gltaA gene sequencing results revealed the presence of two Rickettsia species: R. helvetica and Ca. R. uralica. Rickettsia helvetica DNA was detected in the majority of Rickettsia-positive tick samples (97.1%; 100/103). It was detected in 9.97% (99/993) of I. ricinus and in one of 117 I. trianguliceps. It is noteworthy that the R. helvetica-positive I. trianguliceps was attached to the same animal (My. glareolus) as R. helvetica-positive and -negative I. ricinus (Table 1). Rickettsia helvetica DNA was detected in ticks removed from yellow-necked mice, bank voles and common shrews at all study locations (Table 1).
Another Rickettsia species was identified as Ca. R. uralica. It was detected in three I. trianguliceps ticks removed from two bank voles collected in Pärnumaa and Järvamaa counties, respectively. The total positivity rate of Ca. R. uralica in I. trianguliceps was 2.9% (3/117); Ca. R uralica was not detected in I. ricinus (Tables 1, 2).
To confirm species identity and also to reveal possible nucleotide sequence variability within the detected Rickettsia species, we sequenced the partial ompB genes of 20 samples (all 3 samples with Ca. R. uralica and 17 samples with R. helvetica) and the partial sca4 genes of nine samples (all 3 samples with Ca. R. uralica and 6 samples with R. helvetica). All sequenced R. helvetica partial gene fragments were identical to each other as well as to those previously detected in questing ticks from Estonia [12]. Sequences of gltA, ompB and sca4 gene fragments amplified from all Ca. R. uralica-positive samples were 100% identical to each other; the gltA and sca4 gene fragments were also 100% identical to initial sequences reported from Siberia (Genbank accession numbers KR150785 and KP747665). The ompB gene fragment differed in one nucleotide base, giving 99.9% similarity to the Siberian Ca. R. uralica partial ompB sequence (Genbank accession number KR150780) [7].
Discussion
In this study, ticks of the generalist species I. ricinus and I. persulcatus, as well as nidicolous I. trianguliceps, all attached to small mammals, were analyzed for the presence of vector-borne Rickettsia spp., including a species not previously reported in Europe.
Many studies have focused on the circulation of Rickettsia species in the environment in terms of their vectors, ticks and fleas, as well as in vector-associated mammals, and the possible presence of a Rickettsia reservoir [18, 20, 25,26,27]. In the course of screening vector arthropods and their hosts, an increasing number of new “Candidatus” Rickettsia species have been identified [7, 13]. To date, however, the connection between mammals and rickettsiae has received little attention. Xenodiagnosis studies have shown negative results for R. helvetica [20]. In addition, the percentage of collecting Rickettsia spp. from the positive host is low, as described by Tomassone et al. [21]. Additional studies are required to determine the relationship between rodents and rickettsiae, the bacteremia duration, the distribution and natural cycle of Rickettsia spp. and the association of Rickettsia spp. with different arthropod vectors. Also, further research should aim to identify potential reservoir hosts and determine how Rickettsia spp. are maintained in nature.
To our knowledge, this study is the first to report the detection of a newly described species, Ca. R. uralica, in Europe. In this study, the genospecies was detected only in I. trianguliceps ticks removed from voles, which is in agreement with the first report of Ca. R. uralica from Siberia in which resemblance of Ca. R. uralica to I. trianguliceps was shown [7]. The authors of that study claim that the same Rickettsia variant had been previously detected in Myodes rutilus (northern red-backed voles) and S. araneus, both of which are also present in Estonia. Together with I. trianguliceps ticks, these small mammals might play a role in the circulation of this Rickettsia species in nature. Despite the genetic clustering of this newly described Rickettsia within the spotted-fever group, the pathogenic potential of Ca. R. uralica for domestic and wild mammals, pets or humans remains to be studied.
Although spotted fever rickettsioses are known to be emerging diseases that are spreading across the globe, reports of diseases due to R. helvetica infections in humans are scarce. Serological or molecular tools have been used to detect R. helvetica infection in samples collected from patients with suspected Lyme neuroborreliosis in the Netherlands [10], from those manifesting unexplained fever following a tick bite in France and Italy [11] and in those with rash, febrile illness and meningitis in Sweden [8, 9]. Rickettsia helvetica, a tick-borne rickettsiae species, is also frequently detected in Europe and Asia [2, 28, 29], being reported to be the prevalent Rickettsia species in specific regions, such as Germany [30], Slovakia [31] and Sakhalin Island in Russia [29]. Estonia is also a predominant region in terms prevalence of Rickettsia species, as evidenced by > 95% of all Rickettsia species detected in a questing study [12] and in rodent-attached ticks in the present study being R. helvetica. While there are no clinical reports of illness caused by R. helvetica in Estonia to date, the detection of this tick-borne pathogen (TBP) at positivity rates within tick population similar to the positivity rate of 23.3% for Borrelia burgdorferi (s. l.) (I. Golovljova and J. Geller, personal communications) suggests that R. helvetica should be considered during surveillance for tick-borne diseases in Lyme borreliosis-endemic regions.
Rickettsial DNA was detected in 8.7% of all investigated attached ticks and in 10.0% of I. ricinus, compared to 3.4% in I. trianguliceps. High rates of detection of rickettsial DNA in rodent-attached I. ricinus were also recently reported from Lithuania [26] where 22.6% of individually tested larvae (maximum likelihood estimation 26.5%) were positive for Rickettsia spp.
Several TBPs, such as Anaplasma phagocytophilum [32], Neoehrlichia mikurensis and Babesia microti [33], Francisella tularensis [34] have been detected in nidicolous rodent specialists I. trianguliceps ticks removed from small mammals. As reported by Igolkina et al. [7], SFG Rickettsia was found in 41.2% (14/34) of analyzed I. trianguliceps ticks feeding on voles in Western Siberia, which is significantly higher than the results reported here in our study (3.4%, 4/117). Nevertheless, the role of I. trianguliceps in the circulation and maintenance of TBPs is still largely unknown as is its importance and participation in the transmission of pathogens between ticks and rodent hosts.
In our study, the absence of rickettsial DNA in rodent-attached I. persulcatus larvae (0/64) and nymphs (0/12) could be explained by the relatively small number of I. persulcatus covered in the current study. However, several Rickettsia species, such as Ca. R. tarasevichiae (1/530, 0.2%) and R. helvetica (8/530, 1.5%) were previously reported in unfed questing I. persulcatus ticks in Estonia [12].
We mainly found rickettsial DNA in ticks removed from My. glareolus and A. flavicollis, although it was also presented in some ticks collected from several S. araneus. There are reports of the detection of R. helvetica in various small- to large-sized wild mammal samples from Lithuania [35], the Netherlands and Germany [18, 30, 36] and also in Erithacus rubecula (European robins) and Prunella modularis (dunnocks) from Hungary [37]; however, the significance of these animals in the transmission and maintenance cycle of Rickettsia is still debatable [20]. The Rickettsia spp. infection rates in ticks removed from the same animal varied from 4.8 to 100%, most likely indicating that the ectoparasites might acquire these pathogens not only during blood meals on these animals, but also through previous infections by transstadial, transovarial or horizontal transmission [38]. However, as there were no animal samples tested for the presence of rickettsial DNA in the current study, there is no compelling evidence on whether ticks of this study could have acquired the detected Rickettsia through feeding.
Surprisingly, 42.7% (44/103) of all Rickettsia-positive ticks were removed from rodents caught in Pärnumaa county. Although this region was not covered in the previous study on Rickettsia spp. in questing ticks in Estonia [12], a high rate (28%) of Rickettsia DNA was also detected in questing ticks in Pärnumaa (M. Vikentjeva, J. Geller, I. Golovljova, unpublished observations). Interestingly, this region has previously not shown such high infection rates with any TBP [39,40,41]. However, our longitudinal observations on ticks indicate that the local environment and climate of western coastal Estonia may provide favorable conditions for tick population maintenance and survival, as ticks have always been abundant in these areas (I. Golovljova, unpublished observations).
Conclusion
The results of our study show a higher rate of positivity of Rickettsia spp. in ticks from small mammals compared to ones obtained previously in questing ticks. The high Rickettsia positivity rate in larvae might indicate a transovarial transmission of R. helvetica and the possibility of successful co-feeding transmission while feeding on the same host. Rickettsia helvetica was the most prevalent species and was most frequently detected in I. ricinus ticks, which are considered to be its main vector and the natural reservoir host. This study also provides the first report on the presence of the novel Rickettsia species Ca. R. uralica, initially reported from Siberian regions of Russia, in Estonian populations of I. trianguliceps.
Availability of data and materials
All additional data associated with this study can be obtained from the corresponding author on reasonable request. Unique sequences of Candidatus Rickettsia uralica obtained during this study were submitted to GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MT063090–MT063092.
Abbreviations
- ITS2:
-
Internal transcribed spacer 2
- SFG:
-
Spotted fever group
- TBP:
-
Tick-borne pathogens
References
Merhej V, Raoult D. Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc. 2011;86(2):379–405. https://doi.org/10.1111/j.1469-185X.2010.00151.x.
Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26(4):657–702. https://doi.org/10.1128/CMR.00032-13.
Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10(4):694–719.
Azad AF, Beard CB. Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1998;4(2):179–86. https://doi.org/10.3201/eid0402.980205.
Eremeeva ME, Dasch GA. Challenges posed by tick-borne rickettsiae: eco-epidemiology and public health implications. Front Public Health. 2015;3:55. https://doi.org/10.3389/fpubh.2015.00055.
Dobec M, Golubic D, Punda-Polic V, Kaeppeli F, Sievers M. Rickettsia helvetica in Dermacentor reticulatus ticks. Emerg Infect Dis. 2009;15(1):98–100. https://doi.org/10.3201/eid1501.080815.
Igolkina YP, Rar VA, Yakimenko VV, Malkova MG, Tancev AK, Tikunov AY, et al. Genetic variability of Rickettsia spp. in Ixodes persulcatus/Ixodes trianguliceps sympatric areas from Western Siberia, Russia: identification of a new Candidatus Rickettsia species. Infect Genet Evol. 2015;34:88–93. https://doi.org/10.1016/j.meegid.2015.07.015.
Nilsson K. Septicaemia with Rickettsia helvetica in a patient with acute febrile illness, rash and myasthenia. J Infect. 2009;58(1):79–82. https://doi.org/10.1016/j.jinf.2008.06.005.
Nilsson K, Elfving K, Pahlson C. Rickettsia helvetica in patient with meningitis, Sweden, 2006. Emerg Infect Dis. 2010;16(3):490–2. https://doi.org/10.3201/eid1603.090184.
Koetsveld J, Tijsse-Klasen E, Herremans T, Hovius JW, Sprong H. Serological and molecular evidence for spotted fever group Rickettsia and Borrelia burgdorferi sensu lato co-infections in The Netherlands. Ticks Tick Borne Dis. 2016;7(2):371–7. https://doi.org/10.1016/j.ttbdis.2015.12.010.
Fournier PE, Allombert C, Supputamongkol Y, Caruso G, Brouqui P, Raoult D. Aneruptive fever associated with antibodies to Rickettsia helvetica in Europe and Thailand. J Clin Microbiol. 2004;42(2):816–8.
Katargina O, Geller J, Ivanova A, Varv K, Tefanova V, Vene S, et al. Detection and identification of Rickettsiaspecies inIxodes tick populations from Estonia. Ticks Tick Borne Dis. 2015;6(6):689–94. https://doi.org/10.1016/j.ttbdis.2015.06.001.
Shpynov S, Fournier PE, Rudakov N, Raoult D. “Candidatus Rickettsia tarasevichiae” in Ixodes persulcatus ticks collected in Russia. Ann N Y Acad Sci. 2003;990:162–72. https://doi.org/10.1111/j.1749-6632.2003.tb07358.x.
Yuan TT, Ma L, Jiang BG, Fu WM, Sun Y, Jia N, et al. First confirmed infection of Candidatus Rickettsia tarasevichiae in rodents collected from northeastern China. Vector Borne Zoonotic Dis. 2020;20(2):88–92. https://doi.org/10.1089/vbz.2019.2443.
Wachter M, Wolfel S, Pfeffer M, Dobler G, Kohn B, Moritz A, et al. Serological differentiation of antibodies against Rickettsia helvetica, R. raoultii, R. slovaca, R. monacensis and R. felis in dogs from Germany by a micro-immunofluorescent antibody test. Parasites Vectors. 2015;8:126. https://doi.org/10.1186/s13071-015-0745-1.
Segura F, Pons I, Miret J, Pla J, Ortuno A, Nogueras MM. The role of cats in the eco-epidemiology of spotted fever group diseases. Parasites Vectors. 2014;7:353. https://doi.org/10.1186/1756-3305-7-353.
Walter KS, Carpi G, Evans BR, Caccone A, Diuk-Wasser MA. Vectors as epidemiological sentinels: patterns of within-tick Borrelia burgdorferi diversity. PLoS Pathog. 2016;12(7):e1005759. https://doi.org/10.1371/journal.ppat.1005759.
Sprong H, Wielinga PR, Fonville M, Reusken C, Brandenburg AH, Borgsteede F, et al. Ixodes ricinus ticks are reservoir hosts for Rickettsia helvetica and potentially carry flea-borne Rickettsia species. Parasites Vectors. 2009;2(1):41. https://doi.org/10.1186/1756-3305-2-41.
Scheid P, Speck S, Schwarzenberger R, Litzinger M, Balczun C, Dobler G. Detection of Rickettsia helvetica inIxodes ricinus infesting wild and domestic animals and in a botfly larva (Cephenemyia stimulator) infesting roe deer in Germany. Ticks Tick Borne Dis. 2016;7(6):1268–73. https://doi.org/10.1016/j.ttbdis.2016.07.002.
Burri C, Schumann O, Schumann C, Gern L. Are Apodemus spp. mice and Myodes glareolus reservoirs for Borrelia miyamotoi, Candidatus Neoehrlichia mikurensis, Rickettsia helvetica, R. monacensis and Anaplasma phagocytophilum? Ticks Tick Borne Dis. 2014;5(3):245–51. https://doi.org/10.1016/j.ttbdis.2013.11.007.
Tomassone L, Portillo A, Novakova M, de Sousa R, Oteo JA. Neglected aspects of tick-borne rickettsioses. Parasites Vectors. 2018;11(1):263. https://doi.org/10.1186/s13071-018-2856-y.
Moran-Cadenas F, Schneider H, Lommano E, Burri C, Moret J, Gern L. A comparison of two DNA extraction approaches in the detection of Borrelia burgdorferi sensu lato from live Ixodes ricinus ticks by PCR and reverse line blotting. Vector Borne Zoonotic Dis. 2007;7(4):555–61. https://doi.org/10.1089/vbz.2006.0596.
Varv K, Ivanova A, Geller J, Remm J, Jaik K, Tikunova N, et al. Identification of I. ricinus, I. persulcatus and I. trianguliceps species by multiplex PCR. Ticks Tick Borne Dis. 2017;8(2):235–40. https://doi.org/10.1016/j.ttbdis.2016.11.004.
Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsiausing the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol. 2000;50(Pt 4):1449–55. https://doi.org/10.1099/00207713-50-4-1449.
Minichova L, Hamsikova Z, Mahrikova L, Slovak M, Kocianova E, Kazimirova M, et al. Molecular evidence of Rickettsia spp. in ixodid ticks and rodents in suburban, natural and rural habitats in Slovakia. Parasites Vectors. 2017;10(1):158. https://doi.org/10.1186/s13071-017-2094-8.
Radzijevskaja J, Kaminskiene E, Lipatova I, Mardosaite-Busaitiene D, Balciauskas L, Stanko M, et al. Prevalence and diversity of Rickettsiaspecies in ectoparasites collected from small rodents in Lithuania. Parasites Vectors. 2018;11(1):375. https://doi.org/10.1186/s13071-018-2947-9.
Schex S, Dobler G, Riehm J, Muller J, Essbauer S. Rickettsia spp. in wild small mammals in Lower Bavaria, South-Eastern Germany. Vector Borne Zoonotic Dis. 2011;11(5):493–502. https://doi.org/10.1089/vbz.2010.0060.
Kartashov MY, Glushkova LI, Mikryukova TP, Korabelnikov IV, Egorova YI, Tupota NL, et al. Detection of Rickettsia helvetica and Candidatus R. tarasevichiae DNA in Ixodes persulcatus ticks collected in Northeastern European Russia (Komi Republic). Ticks Tick Borne Dis. 2017;8(4):588–92. https://doi.org/10.1016/j.ttbdis.2017.04.001.
Igolkina Y, Bondarenko E, Rar V, Epikhina T, Vysochina N, Pukhovskaya N, et al. Genetic variability of Rickettsia spp. in Ixodes persulcatus ticks from continental and island areas of the Russian Far East. Ticks Tick Borne Dis. 2016;7(6):1284–9. https://doi.org/10.1016/j.ttbdis.2016.06.005.
Fischer S, Spierling NG, Heuser E, Kling C, Schmidt S, Rosenfeld UM, et al. High prevalence of Rickettsia helvetica in wild small mammal populations in Germany. Ticks Tick Borne Dis. 2018;9(3):500–5. https://doi.org/10.1016/j.ttbdis.2018.01.009.
Svehlova A, Berthova L, Sallay B, Boldis V, Sparagano OA, Spitalska E. Sympatric occurrence of Ixodes ricinus, Dermacentor reticulatus and Haemaphysalis concinna ticks and Rickettsia and Babesia species in Slovakia. Ticks Tick Borne Dis. 2014;5(5):600–5. https://doi.org/10.1016/j.ttbdis.2014.04.010.
Rar VA, Epikhina TI, Yakimenko VV, Malkova MG, Tancev AK, Bondarenko EI, et al. Genetic variability of Anaplasma phagocytophilum in ticks and voles from Ixodes persulcatus/Ixodes trianguliceps sympatric areas from Western Siberia, Russia. Ticks Tick Borne Dis. 2014;5(6):854–63. https://doi.org/10.1016/j.ttbdis.2014.07.008.
Blanarova L, Stanko M, Miklisova D, Vichova B, Mosansky L, Kraljik J, et al. Presence of Candidatus Neoehrlichia mikurensis and Babesia microti in rodents and two tick species (Ixodes ricinusandIxodes trianguliceps) in Slovakia. Ticks Tick Borne Dis. 2016;7(2):319–26. https://doi.org/10.1016/j.ttbdis.2015.11.008.
Kormilitsyna MI, Korenberg EI, Kovalevskii YV, Meshcherakova IS. First molecular identification of the Tularemia agent in the ticks Ixodes trianguliceps Bir. in Russia. Mol Gen Mikrobiol Virusol. 2016;34(2):67–70.
Mardosaite-Busaitiene D, Radzijevskaja J, Balciauskas L, Paulauskas A. First detection of Rickettsia helvetica in small mammals in Lithuania. New Microbes New Infect. 2018;22:19–23. https://doi.org/10.1016/j.nmni.2017.12.012.
Obiegala A, Oltersdorf C, Silaghi C, Kiefer D, Kiefer M, Woll D, et al. Rickettsia spp. in small mammals and their parasitizing ectoparasites from Saxony, Germany. Vet Parasitol Reg Stud Rep. 2016;5:19–24. https://doi.org/10.1016/j.vprsr.2016.08.008.
Hornok S, Kovats D, Csorgo T, Meli ML, Gonczi E, Hadnagy Z, et al. Birds as potential reservoirs of tick-borne pathogens: first evidence of bacteraemia with Rickettsia helvetica. Parasites Vectors. 2014;7:128. https://doi.org/10.1186/1756-3305-7-128.
Socolovschi C, Mediannikov O, Raoult D, Parola P. The relationship between spotted fever group Rickettsiae and ixodid ticks. Vet Res. 2009;40(2):34. https://doi.org/10.1051/vetres/2009017.
Geller J, Nazarova L, Katargina O, Golovljova I. Borrelia burgdorferi sensu lato prevalence in tick populations in Estonia. Parasites Vectors. 2013;6:202. https://doi.org/10.1186/1756-3305-6-202.
Katargina O, Geller J, Alekseev A, Dubinina H, Efremova G, Mishaeva N, et al. Identification of Anaplasma phagocytophilum in tick populations in Estonia, the European part of Russia and Belarus. Clin Microbiol Infect. 2012;18(1):40–6. https://doi.org/10.1111/j.1469-0691.2010.03457.x.
Katargina O, Russakova S, Geller J, Kondrusik M, Zajkowska J, Zygutiene M, et al. Detection and characterization of tick-borne encephalitis virus in Baltic countries and eastern Poland. PLoS One. 2013;8(5):e61374. https://doi.org/10.1371/journal.pone.0061374.
Acknowledgements
The authors would like to thank Kertu Jaik (University of Tartu, Estonia) and Larissa Starkova (National Institute for Health Development, Estonia), for their invaluable assistance in the animal and laboratory experiments. The authors also thank Sirkka Vene for helpful advice and suggestions.
Funding
The current study was financially supported by the program TerVe, project ZoonRisk 3.2.1002.11-0002 (European Regional Development Fund, Estonian Research Council), and the project IUT-42-1 (Ministry of Education and Research, Estonia).
Author information
Authors and Affiliations
Contributions
JR and IG study initiated and planned the experimental studies; JR collected samples and identified the small mammal species identification; MV and JG performed DNA extraction, PCR analyses and the bioinformatics analysis; MV wrote the initial draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were approved by Estonian Ministry of Agriculture Permission No. 124 (J. Remm).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Rickettsia spp. in ticks from infested small mammal species.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Vikentjeva, M., Geller, J., Remm, J. et al. Rickettsia spp. in rodent-attached ticks in Estonia and first evidence of spotted fever group Rickettsia species Candidatus Rickettsia uralica in Europe. Parasites Vectors 14, 65 (2021). https://doi.org/10.1186/s13071-020-04564-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04564-7