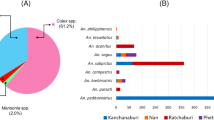
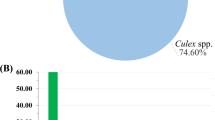
Abstract
Background
Plasmodium knowlesi is a significant cause of human malaria in Sarawak, Malaysian Borneo. Only one study has been previously undertaken in Sarawak to identify vectors of P. knowlesi, where Anopheles latens was incriminated as the vector in Kapit, central Sarawak. A study was therefore undertaken to identify malaria vectors in a different location in Sarawak.
Methods
Mosquitoes found landing on humans and resting on leaves over a 5-day period at two sites in the Lawas District of northern Sarawak were collected and identified. DNA samples extracted from salivary glands of Anopheles mosquitoes were subjected to nested PCR malaria-detection assays. The small subunit ribosomal RNA (SSU rRNA) gene of Plasmodium was sequenced, and the internal transcribed spacer 2 (ITS2) and mitochondrial cytochrome c oxidase subunit 1 (cox1) gene of the mosquitoes were sequenced from the Plasmodium-positive samples for phylogenetic analysis.
Results
Totals of 65 anophelines and 127 culicines were collected. By PCR, 6 An. balabacensis and 5 An. donaldi were found to have single P. knowlesi infections while 3 other An. balabacensis had either single, double or triple infections with P. inui, P. fieldi, P. cynomolgi and P. knowlesi. Phylogenetic analysis of the Plasmodium SSU rRNA gene confirmed 3 An. donaldi and 3 An. balabacensis with single P. knowlesi infections, while 3 other An. balabacensis had two or more Plasmodium species of P. inui, P. knowlesi, P. cynomolgi and some species of Plasmodium that could not be conclusively identified. Phylogenies inferred from the ITS2 and/or cox1 sequences of An. balabacensis and An. donaldi indicate that they are genetically indistinguishable from An. balabacensis and An. donaldi, respectively, found in Sabah, Malaysian Borneo.
Conclusions
Previously An. latens was identified as the vector for P. knowlesi in Kapit, central Sarawak, Malaysian Borneo, and now An. balabacensis and An. donaldi have been incriminated as vectors for zoonotic malaria in Lawas, northern Sarawak.

Similar content being viewed by others
Background
Human infections with Plasmodium knowlesi, a malaria parasite of long-tailed and pig-tailed macaques [1], were thought to be extremely rare until a large number of human cases were described in the Kapit Division of Sarawak State, Malaysian Borneo [2]. Knowlesi malaria cases have been reported from different locations in Malaysia [2,3,4,5,6,7], in almost all countries in Southeast Asia [8,9,10,11,12,13,14,15,16,17] except Timor Leste, and also in the Nicobar and Andaman Islands of India [18]. This simian malaria parasite is now the most common cause for malaria admissions to hospitals in Malaysia, where in the years 2017–2019, 10,968 cases were reported, with 87% occurring in the states of Sabah and Sarawak in Malaysian Borneo (unpublished data, Ministry of Health Malaysia) [19]. Despite now being recognised as a significant cause of malaria in humans and an additional challenge to malaria elimination in Southeast Asia [20,21,22,23], there remains a paucity of information regarding the vectors of P. knowlesi in nature.
In the period between 1951 and 1997, malaria vectors in Sarawak, Malaysian Borneo were determined by examining dissected salivary glands for the presence of sporozoites. Information about vectors of human malaria parasites was first reported by McArthur [24] in his 1951 review regarding the importance of the Leucosphyrus Group of Anopheles in malaria transmission. Dissection of various species of Anopheles mosquitoes was carried out in the Kuching General Hospital and oocysts were found in the midguts of An. latens. Later, in 1956 and 1995, An. latens, An. barbirostris (s.l.) and An. donaldi were incriminated as the vectors for human malaria parasites in Upper Baram [25, 26]. In Miri, Chang et al. [27] discovered malaria sporozoites in the salivary glands of An. donaldi and An. letifer in an oil palm plantation in Miri in 1997. However, the sporozoites of Plasmodium spp. harboured by these vectors could not be determined accurately since molecular tools were not available when these studies were undertaken. Despite this, intravenous inoculation of sporozoites isolated from the simiophilic An. hackeri in 1961 into an uninfected rhesus monkey demonstrated that An. hackeri was the natural vector of P. knowlesi in Peninsular Malaysia [28]. The parasite was morphologically identified from the blood of the rhesus monkey inoculated with the sporozoites. It was only in 1999, in Belaga, Sarawak that an ELISA was used for detection of sporozoites of Plasmodium spp. in mosquitoes, but the ELISA could only detect two species of Plasmodium, P. falciparum and P. vivax [29].
The first and only entomological study in Sarawak, Malaysian Borneo that identified Plasmodium species using molecular methods was conducted in 2005–2006. Using nested PCR assays on DNA extracted from salivary glands of mosquitoes in the Kapit District, An. latens was incriminated as the vector of P. knowlesi in this region [30]. Subsequently, similar studies utilising molecular tools have incriminated An. cracens as the vector of P. knowlesi in Kuala Lipis, Peninsular Malaysia [31] while An. balabacensis was found to harbour sporozoites of P. coatneyi, P. cynomolgi, P. inui, P. knowlesi, and an unidentified Plasmodium species in Sabah, Malaysian Borneo [32]. Anopheles introlatus was hypothesised to be a vector in Selangor, Peninsular Malaysia, as Vythilingam et al. [33] discovered one out of 55 An. introlatus to be infected with only oocysts, but not sporozoites of P. knowlesi. In southern Vietnam where co-infection of P. vivax and P. knowlesi was predominant in humans and mosquitoes, An. dirus was shown to be the vector of P. falciparum, P. vivax and P. knowlesi using molecular methods [15]. Information derived from these studies was undeniably imperative for vector control but similar knowledge of vectors of P. knowlesi is lacking due to restricted sampling only in the Kapit District in Sarawak [28].
Molecular studies of malaria vectors have indicated that morphological keys cannot differentiate sibling species within a species complex and have revealed extensive cryptic speciation, with most nominal species in Southeast Asia being found to comprise species complexes [34,35,36]. The Dirus Complex alone, for example, consists of at least eight species [37, 38]. For the purpose of accurate identification, DNA barcodes such as the second internal transcribed spacer (ITS2) within the ribosomal DNA, mitochondrial cytochrome c oxidase subunits 1 and 2 (cox1 and cox2), and NADH dehydrogenase subunit 6 (nad6) were previously used to study the phylogeny of closely-related mosquito species [35, 39,40,41,42,43,44,45]. Accurate identification of vectors is especially important to inform malaria vector control programmes in Southeast Asia, where most Anopheles malaria vectors are comprised of species complexes [23, 36], and where malaria elimination is on the agenda [46]. It is therefore critical to utilise available molecular methods to precisely identify Anopheles mosquitoes found to be vectors for zoonotic malaria parasites.
The present study was aimed at incriminating the vector(s) of P. knowlesi and other malaria parasites in northern Sarawak (Lawas District), as well as providing an accurate identity of the incriminated vector(s). Lawas District was selected as the study site since 173 patients with knowlesi malaria had been admitted to the Lawas Hospital three years prior to the commencement of the study in 2014 (A. Ting, Lawas Hospital, personal communication).
Methods
Study site
The study was carried out in the Lawas District of northern Sarawak, Malaysian Borneo (Fig. 1). Lawas District is bordered by Brunei on its west, Sabah on its east, North Kalimantan on its south, and Labuan Federal Territory on its north. The first mosquito collection site was close to Long Tengoa village (DMS: 4° 37′ 5′′ N, 115° 20′ 23′′ E) which had 3 recent human cases of knowlesi malaria prior to mosquito collection in September 2014. The collection site is situated in a forested area c. 500 m eastward from the village at an elevation of about 100 m above sea level. The second collection was conducted in an abandoned army camp (4° 16′ 18′′ N, 115° 31′ 49′′ E) close to Long Luping, approximately 40 km southward from Long Tengoa. This camp is situated at an elevation of c. 650 m above sea level right beside a stream. The three patients who were admitted to Lawas Hospital with knowlesi malaria within three months prior to mosquito collection in May 2015 had spent time hunting near this camp site.
Locations of entomological surveys conducted previously and for the present study [74]. Human malaria sporozoites were discovered from dissected mosquitoes from these sites between 1951–1999 [24,25,26,27, 29] (triangles). Plasmodium knowlesi and other simian malaria sporozoites were discovered from dissected mosquitoes and by nested PCR assays from this site between 2005–2006 [30] (circle). The study site for the present study is indicated by a square
Mosquito collection, identification and dissection
The first mosquito collection was carried out by 6 collectors for a period of 3 days in September 2014 in Long Tengoa village. The second collection at Long Luping was carried out by 5 collectors for 2 days in May 2015. All mosquitoes collected in Long Tengoa (18:00–23:00 h) were collected using the human landing catch method while those collected in Long Luping (day 1: 16:00–22:00 h; Day 2: 16:00–21:00 h) were collected using both the human landing catch method and the resting catch method. For the human landing catch method, all mosquitoes landing/biting on human bait were caught using a cylindrical specimen tube (18 mm in diameter × 50 mm in height) with moist tissue covering the bottom of the tube. Once a mosquito was caught, the opening of the tube was plugged with cotton wool and labelled according to the time of collection. The procedures for resting catch method were similar to that of the human landing catch method except that mosquitoes found resting under leaves were collected. The mosquitoes were then brought to the field laboratory for morphological identification. Anopheles mosquitoes were identified to the species/group level using taxonomic keys while non-anophelines were identified to the generic level [47,48,49]. Salivary glands of individual Anopheles mosquitoes were then carefully dissected and preserved in 1.5-ml microcentrifuge tubes (1 specimen/tube) containing 0.5 ml absolute ethanol. The dissection pins were wiped with dispensable 70% ethanol swabs after every dissection to prevent cross-contamination. Preserved salivary glands were transported to the Malaria Research Centre, Universiti Malaysia Sarawak, for further molecular analysis.
DNA extraction and detection of Plasmodium species
Absolute ethanol preserving the salivary glands was first dried prior to DNA extraction. Genomic DNA of the dried salivary glands was extracted using DNeasy® Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The eluted samples were kept at 4 °C until required. Extracted DNA samples were initially subjected to nested PCR assays for the detection of Plasmodium DNA based on the SSU rRNA genes using the Plasmodium-specific primers rPLU3 and rPLU4 [50]. Subsequently, Plasmodium-positive samples were tested using a species-specific PCR assays to identify P. coatneyi, P. cynomolgi and P. knowlesi [1]. Additionally, new PCR primers were developed for P. fieldi (PfldF3: 5′-GAT CTT TTT TTG TTT CGG CAT TGA A-3′; PfldR3: 5′-AAG GCA CTG AAG GAA GCA ATC TAA GAG TTT-3′) and P. inui (PinF5: 5′-GTA TCG ACT TTG TGC GCA TTT TTC TAC-3′; INAR3: 5′-GCA ATC TAA GAG TTT TAA CTC CTC-3′) with optimum annealing temperatures at 60 °C and 62 °C, respectively. The new primers were found to have higher specificity (data not shown) compared to the P. fieldi-specific and P. inui-specific primers developed previously [1]. The processes of genomic DNA extraction, PCR mastermix preparation, pipetting of template for primary PCR, and pipetting of template for nested PCR were each conducted in a different room, using filtered pipette tips and micropipettes dedicated to each room to prevent cross-contamination. These PCR assays were repeated for samples that were positive for the initial nested PCR assays and only samples which were consistently positive on both occasions were subjected to sequencing of Plasmodium SSU rRNA genes.
Generating Plasmodium SSU rRNA gene amplicons
The SSU rRNA gene fragments were amplified by semi-nested PCR assays in order to increase the DNA yield prior to cloning. Nest 1 PCR amplification was performed using primers rPLU1 and rPLU5 as previously described [50]. Then, PCR product from Nest 1 was used as DNA template for the Nest 2 PCR amplification using the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, USA). PCR primers rPLU3 and rPLU5 [50] were used with an annealing temperature of 68 °C.
Generating Anopheles cox1 and ITS2 region amplicons
Both the cox1 and ITS2 regions of the vectors were amplified by PCR assays using the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) according to the manufacturer’s protocol. To amplify a 748 bp-long cox1 region, PCR primers AnCOX1F (5′-GGA ATG GAT GTW GAT ACW CGA GC-3′) and AnCOX1R (5′-CCT AAA TTT GCT CAT GTT GCC-3′) were designed with the annealing temperature of 65 °C. For the ITS2 region, DNA amplification was performed using the PCR primers 5.8SF and 28SR as previously described by Paredes-Esquivel et al. [35].
Cloning and sequencing
Generated amplicons were ligated with pCR®-Blunt Vector (Invitrogen, Carlsbad, USA) and transformed into One Shot® TOP10 Chemically Competent E. coli (Invitrogen), according to the Zero Blunt® PCR Cloning Kit (Invitrogen) protocol. Escherichia coli transformed colonies were screened with PCR using M13 primers, for the presence of target DNA insert and plasmids containing the inserts were purified using the PureLink® Quick Plasmid Miniprep Kit (Thermo Fisher Scientific). DNA sequencing of plasmid DNA was conducted according to the BigDye® Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) protocol, using M13 primers, internal primers rPLU2 and rPLU6 for the Plasmodium SSU rRNA inserts, and internal primers JIBF (5′-CTA GTG TGC TTC CCA TGG AGA TAG-3′) and JIBR (5′-CAC ACC CAA CCC AAT AAA AAT TG-3′) for ITS2 inserts.
Phylogenetic analysis
For the ITS2 region, the central repeat region contained within it was identified using the Tandem Repeats Finder software (https://tandem.bu.edu/trf/trf.html) as described previously and trimmed [35, 51]. The trimmed sequences were then aligned for phylogenetic analysis. No trimming was done on the SSU rDNA and cox1 sequences prior to alignment. Reference sequences obtained from GenBank [35, 40, 52,53,54] are listed in Additional file 1: Table S1 and Additional file 2: Table S2. Multiple sequence alignments were performed using the default parameters of ClustalW within the LaserGene 7.1 programme (DNASTAR; Lasergene, Inc., Madison, WI, USA). The best nucleotide substitution models were calculated using MEGA 7.0.21 and the models with the lowest Bayesian information criterion (BIC) were selected [55, 56]. Subsequently, phylogenetic trees were constructed by the Maximum Likelihood (ML) method using MEGA 7.0.21 with bootstrap values calculated from 1000 replicates [57].
Results
Species composition of collected mosquitoes
A total of 192 mosquitoes were collected from Long Tengoa (n = 86) and Long Luping (n = 106) (Table 1). Based on morphological characteristics, mosquitoes of the genus Anopheles were the most abundant (n = 65; 34%), followed by Culex spp. (n = 41; 21%), Aedes spp. (n = 40; 21%), Armigeres spp. (n = 31; 16%), Mansonia spp. (n = 7; 4%), Malaya spp. (n = 6; 3%) and Uranotaenia spp. (n = 2; 1%).
A higher proportion of culicines was found in Long Tengoa (n = 72; 84%), while Anopheles mosquitoes predominated in Long Luping (n = 51; 48%). Most An. balabacensis (n = 31) and An. barbirostris (s.l.) (n = 18) mosquitoes were caught at Long Luping while all specimens of the Umbrosus Group (An. letifer, An. roperi and An. umbrosus) were collected in Long Tengoa, and these were not collected in Long Luping (Table 1).
Plasmodium species in Anopheles mosquitoes
DNA extracted from the salivary glands of 65 Anopheles mosquitoes was subjected to nested PCR assays and Plasmodium DNA was only detected in samples derived from 10 An. balabacensis and 6 An. barbirostris (s.l.) collected in Long Luping. Of 31 An. balabacensis, 9 (29.0 %) were found to carry P. knowlesi and other simian malaria parasites while 5 (27.8 %) An. barbirostris (s.l.) were found to be infected with P. knowlesi only (Table 2). The identity of the species of Plasmodium could not be determined for 2 of the Plasmodium-positive samples by using the species-specific PCR primers for P. coatneyi, P. cynomolgi, P. inui, P. fieldi and P. knowlesi.
All 14 samples which tested positive for at least one simian malaria parasite were subjected to another round of semi-nested PCR to obtain a longer fragment of the SSU rRNA for sequencing. Amplicons could not be generated for 5 (3 An. balabacensis and 2 An. barbirostris (s.l.)) of the samples infected with P. knowlesi only. For the 9 samples where amplicons were successfully obtained, cloned and transformed into chemically competent E. coli, colony PCR was conducted on at least 10 colonies. Colonies originating from the same transformation which produced amplicons of slightly different sizes (1500–2000 bp) were subjected to sequencing. As a result, a total of 38 SSU rRNA fragments were cloned and sequenced from these Anopheles mosquitoes (6 An. balabacensis and 3 An. barbirostris (s.l.) which were Plasmodium-positive by nested PCR assays. The phylogenetic tree (Fig. 2) constructed using the Plasmodium A-type and S-type SSU rRNA sequences showed that An. balabacensis were harbouring sporozoites of P. cynomolgi (LW67), P. inui (LW67 and LW74) and P. knowlesi (LW45, LW59, LW31 and LW49) in their salivary glands, while An. barbirostris (s.l.) carried only P. knowlesi (LW47, LW57 and LW58). Three An. balabacensis (LW45, LW67 and LW74) were shown to carry more than 1 species of Plasmodium while the other 2 An. balabacensis (LW31 and LW49) were infected with P. knowlesi only. Multiple SSU rRNA sequences derived from the salivary glands of mosquitoes LW45, LW67 and LW74 formed several distinct clades instead of grouping with any of the clades formed by the reference sequences (Fig. 2). This indicates that An. balabacensis is not only harbouring sporozoites of P. cynomolgi, P. inui and P. knowlesi, but probably sporozoites of unknown species of Plasmodium.
Phylogenetic tree for Plasmodium spp. based on the SSU rRNA genes using the ML method. The analysis was based in the Tamura 3-parameter + G + I substitution model. GenBank accession numbers are given in parentheses; letters ‘A’ and ‘S’ represent the different isoforms of the SSU rRNA gene. Only bootstrap values > 70% are shown at the nodes. Blue and red colours represent parasites obtained from An. barbirostris (s.l.) and An. balabacensis, respectively
Despite detecting P. cynomolgi and P. fieldi by PCR in LW45 and LW67, respectively, we were unable to obtain SSU rRNA sequences of these two species of Plasmodium from the clones that were sequenced (Fig. 2). Binding sites for P. cynomolgi–specific and -P. fieldi-specific primers were subsequently searched for in sequences isolated from LW45 and LW67 to confirm their specificity in detecting the species they were developed to identify. Clones LW45C10 and LW45C13 were found to have fully complementary binding sites for the P. cynomolgi-specific primers (CY2F + CY4R) although they did not form a clade with P. cynomolgi in the phylogenetic tree (Fig. 2). CY2F binding site was also discovered to be fully conserved in several other clones isolated from LW67 (C3, C10, C14, C20 and C30) while one and/or two SNPs were present for primer CY4R (Fig. 3). As the SNPs were not near to the 3′-end of the primer binding site, CY2F + CY4R could potentially amplify a 137-bp fragment from these aligned clones in a PCR. On the other hand, P. fieldi-specific primer sequences were not conserved in any of the clones isolated from LW67. This suggests that the P. fieldi detected by PCR was not recovered among the 23 clones sequenced.
Molecular characterisation of Anopheles mosquitoes
The ITS2 region and cox1 gene (for multiple sequence alignment of cox1 see Additional file 3: Alignment S1, Additional file 4: Alignment S2) of Anopheles mosquitoes carrying simian malaria parasites were further sequenced to confirm their identities. Phylogenetic analysis of the ITS2 region and cox1 gene of An. barbirostris (s.l.) from Thailand, West Sumatra, West Java, South Kalimantan, and Sabah showed 6 distinct clades (Figs. 4, 5). The phylogenetic trees constructed using the ML method showed that sequences derived from An. barbirostris (s.l.) collected from the Lawas District, Sarawak, clustered together with An. donaldi (Figs. 4, 5) from Sabah and An. barbirostris (s.l.) (Fig. 4) from Selangor, Peninsular Malaysia. This indicates that the P. knowlesi-positive An. barbirostris (s.l.) collected in the present study are An. donaldi.
Phylogenetic tree for Anopheles spp. based on the ITS2 region using the ML method. The analysis was based in the Kimura 2-parameter + G substitution model. Only bootstrap values > 70% are shown on the nodes. GenBank accession numbers are given in parentheses. Blue colour represents An. barbirostris (s.l.) collected in Lawas District
Phylogenetic tree for Anopheles spp. based on the cox1 gene using the ML method. The analysis was based in the Tamura 3-parameter + G + I substitution model. Only bootstrap values > 70% are shown on the nodes. GenBank accession numbers are given parentheses. Blue colour represents An. barbirostris (s.l.) collected in Lawas District
The cox1 sequences of An. balabacensis obtained in this study showed close phylogenetic relationships (Fig. 6) with the other An. balabacensis sequences. Those collected from this study in Lawas District, Sarawak, clustered together with the An. balabacensis in eastern Sabah, Malaysian Borneo (GenBank: DQ897940), forming a sister clade to the An. balabacensis from South Kalimantan, Indonesian Borneo (GenBank: DQ897941).
Phylogenetic tree for Anopheles spp. based on the cox1 gene using the ML method. The analysis was based in the Tamura 3-parameter + G substitution model. Only bootstrap values > 70% are shown on the nodes. GenBank accession number are given in parentheses. Red colour represents An. balabacensis collected in Lawas District
Discussion
Identification of Plasmodium
Sporozoites of P. knowlesi and other simian malaria parasites were identified by nested PCR assays in An. balabacensis (n = 9) and An. barbirostris (s.l.) (n = 5) (subsequently identified following molecular characterisation as An. donaldi). The specimens of An. balabacensis and An. donaldi examined had sporozoite infection rates of 29.0% and 27.8% respectively, which are exceedingly high compared to other vector studies in Malaysia. In the neighbouring Malaysian Borneo state of Sabah, An. balabacensis was identified as the vector for P. knowlesi with sporozoite rates ranging from 1.03 to 3.42% at three different sites [32]. Infection rates of < 2.00% were reported among vectors in central Sarawak and among An. cracens in Peninsular Malaysia [29, 31, 58]. The only other study known to have found a comparable sporozoite rate was a study done in Palawan, Philippines, where 29.4% of the examined An. balabacensis had sprorozoites [59]. The high sporozoite rates of the vectors could be attributed to the site of collection as well, which is consistent with the finding of the previous study involving An. latens in Kapit, Sarawak [58], where it was found that the sporozoite rate was highest in the forest, followed by the rates in the farm at the forest-fringe, and in the long house. Long Luping is an abandoned army camp which is far away from any human settlement. It is potentially a foraging site for the macaques due to the banana trees that grow around its perimeter. No vector control activities have been carried out since the army camp was abandoned and macaques were sighted at the site during the entomological surveys, suggesting that macaques and mosquitoes could forage and breed, respectively, undisturbed in this site.
The high infection rates could also be due to the way sampling was conducted compared to other reported studies [4, 30,31,32, 60, 61]. First, the present method recovers both sides of the salivary glands without rupturing any to check for sporozoites, increasing the yield of extracted Plasmodium DNA. Secondly, the present method does not retain the head of the specimens which might contain inhibitors which would disrupt PCR assays. PCR inhibitors were previously found to be present in the heads of Culex pipiens and An. punctipennis which caused false negative results in detection of Wolbachia pipientis in samples tested [62]. The efficiency of the PCR assay was later restored when specimens were decapitated prior to DNA extraction. Although we had high sporozoite infection rates by nested PCR assays, we failed to generate the longer (1500–2000 bp) Plasmodium SSU rDNA amplicons from five samples that were malaria-positive by nested PCR assays. This could be caused by the very low number of sporozoites present in the salivary glands of the mosquitoes and the low sensitivity of the PCR assay to amplify long fragments from a sample with low template concentration.
Diversity and density of Plasmodium infection in vectors
In line with the discovery of at least seven Plasmodium species infecting long-tailed macaques in Sarawak [63], it is unsurprising that multiple unidentified Plasmodium species were recovered from An. balabacensis in this study (Fig. 2). However, the low quantity of DNA extracted from the salivary glands prevented the sequencing of another gene, such as the mitochondrial genome, which would have been necessary to determine whether these are indeed novel species of Plasmodium. It is highly likely that unidentified Plasmodium species co-infected the vectors when they fed on macaques which are known to host a diverse range of species of Plasmodium [1, 4, 63, 64].
Accurate identification of Plasmodium by PCR might also be impeded when uncharacterised Plasmodium, such as those found in sample LW45 (C10 and C13), could be detected by P. cynomolgi-specific primers (Fig. 3). This demonstrates the need for the sequencing of a considerable genomic locus length for proper identification of the species of Plasmodium. On the other hand, the unsuccessful attempts to sequence P. fieldi from sample LW67, and P. cynomolgi from LW45, could be due to the low density of P. fieldi and P. cynomolgi, respectively, among the other Plasmodium co-infecting each of these mosquitoes. As the PCR amplification prior to cloning amplifies the SSU rRNA genes of all Plasmodium species indiscriminately, the scarcity of any species of Plasmodium DNA in the sample reduces the chance of its amplicon being produced during PCR amplification. The difficulty in obtaining sequencing data of genes of Plasmodium derived from vectors is probably the main reason why previous studies on vectors of knowlesi malaria have only used nested PCR assays [15, 30, 32, 33, 61] and for the paucity of studies describing the diversity and density of Plasmodium infection in vectors [31, 60]. With an increasing number of zoonotic malaria infections worldwide [65,66,67], epidemiological studies of these inadequately studied species within human populations that come into close contact with macaques during activities in the forest and forest-fringe will also be required to monitor potential host-switch events.
Molecular characterisation of vectors and implications for vector control in Sarawak
Phylogenetic analyses confirmed that the Plasmodium-positive mosquitoes from Lawas, Sarawak, identified morphologically as An. balabacensis were An. balabacensis whereas those identified as An. barbirostris (s.l.) were An. donaldi. Anopheles balabacensis has been incriminated as a vector for P. knowlesi in Sabah, Malaysian Borneo and belongs to the Leucosphyrus Group which has been long thought to be the only species group capable of transmitting P. knowlesi in natural settings [4, 15, 30,31,32, 60, 61]. Anopheles kochi from the Kochi Group was suspected as a vector due to its high susceptibility to P. knowlesi infection under experimental conditions and its simiophilic biting behaviour but the parasite was never recovered from any An. kochi collected in the natural environment [4, 68, 69]. DNA of P. knowlesi was detected by PCR assays in 2019 from DNA extracted from the carcasses of An. donaldi in Sabah, Malaysian Borneo and from An. sundaicus in the Nicobar and Andaman Islands of India, [61, 70] whereas the Plasmodium DNA detected in the present study in Sarawak, Malaysian Borneo were recovered from the salivary glands of mosquitoes. In both Sabah and Sarawak, more detailed studies need to be conducted on the bionomics of An. donaldi and An. balabacensis to provide data for the implementation of appropriate vector control. Species-specific molecular assays should also be designed and utilised in future vector incrimination studies for this species range in order to correctly identify malaria-infective mosquitoes.
The incrimination of An. balabacensis and An. donaldi as novel vectors for P. knowlesi in northern Sarawak calls for re-evaluation of current and future vector control methods in the state. Detailed studies first need to be undertaken to determine the feeding behaviour and host preference of these vectors. The main vector control methods currently adopted by the Sarawak State Health Department for malaria control are the provision of insecticide treated bednets and the regular spraying of residual insecticide on houses in malarious areas. These would be of limited value against An. latens in central Sarawak which are mainly exophagic and acrodendrophilic [58]. Further studies need to be undertaken on An. donaldi and An. balabacensis in Lawas to determine the effectiveness of providing bednets to people. From the aspect of insecticide-based preventative measures, future insecticide resistance surveys should include both An. balabacensis and An. donaldi to ensure that the insecticide used would still be efficient in killing these vectors. As one of the main vector control methods currently adopted by the Sarawak State Health Department is the regular spraying of residual insecticide on houses in malarious area, spraying could also be considered for uninhabited buildings like the army camp in Long Luping, where human presence is intermittent. Apart from insecticides, clustered regularly interspaced short palindromic repeat (CRISPR)-based gene drive has been recently suggested as one possible method for the control of P. knowlesi vector(s) [71]. Gene drive is a biotechnology method used to increase the spread of a genetic trait (e.g. mosquito sterility/mosquito immunity against Plasmodium infection) into the wild population. As the system relies on the ability of the nuclease to recognise a specific nucleotide sequence and cause a double-stranded break on the chromosome [72], it is imperative that the genomes of the vectors of P. knowlesi are readily available for researchers in the field. Besides An. dirus which is a member of the Leucosphyrus Group, no other An. leucosphyrus (s.l.) and An. barbirostris (s.l.) genomes have been sequenced [73]. Genomic data of vectors of P. knowlesi incriminated so far in Peninsular Malaysia, Malaysian Borneo and Vietnam will be crucial in the development of a gene drive since it is an extremely species-specific vector control method [72]. Novel methods of vector control are clearly needed, and while waiting for these to be developed, personal protection and avoidance of being bitten by mosquitoes need to be advocated during public health promotion exercises as methods for the prevention and control of zoonotic malaria.
Conclusions
The malaria-positive An. balabacensis in Lawas were phylogenetically indistinguishable from An. balabacensis in eastern Sabah, Malaysian Borneo and south Kalimantan, Indonesian Borneo. Phylogenetic analyses also indicated that the Plasmodium-positive mosquitoes identified morphologically as An. barbirostris (s.l.) in Lawas are An. donaldi. Anopheles balabacensis has been incriminated as a vector of P. knowlesi and other simian malaria parasites, and An. donaldi as a vector of P. knowlesi in Lawas, northern Sarawak in Malaysian Borneo. Previously An. latens had been incriminated as the vector for P. knowlesi in Kapit, central Sarawak, so these two species represent novel vectors for zoonotic malaria in Sarawak.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files. Sequences were deposited in GenBank under the accession numbers listed in Additional files 1 and 2.
Abbreviations
- SSU rRNA:
-
Small subunit ribosomal RNA
- cox1:
-
Cytochrome c oxidase subunit 1 gene
- ITS2:
-
Internal transcribed spacer 2 region
- DNA:
-
Deoxyribonucleic acid
- PCR:
-
Polymerase chain reaction
References
Lee KS, Divis PCS, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7:4.
Singh B, Sung LK, Matusop A, Radhakrishnan A, Shamsul SSG, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24.
Cox-Singh J, Davis TME, Lee K, Shamsul SSG, Matusop A, Ratnam S, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–71.
Vythilingam I, Noorazian YM, Huat TC, Jiram AI, Yusri YM, Azahari AH, et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit Vectors. 2008;1:26.
Barber BE, William T, Jikal M, Jilip J, Dhararaj P, Menon J, et al. Plasmodium knowlesi malaria in children. Emerg Infect Dis. 2011;17:814–20.
William T, Menon J, Rajahram G, Chan L, Ma G, Donaldson S, et al. Severe Plasmodium knowlesi malaria in a tertiary care hospital, Sabah. Malaysia. Emerg Infect Dis. 2011;17:7.
Cooper DJ, Rajahram GS, William T, Jelip J, Mohammad R, Benedict J, et al. Plasmodium knowlesi malaria in Sabah, Malaysia, 2015–2017: ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin Infect Dis. 2019;70:361–7.
Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human. Thailand. Emerg Infect Dis. 2004;10:2211–3.
Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, Cui L, et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis. 2009;199:1143–50.
Luchavez J, Espino F, Curameng P, Espina R, Bell D, Chiodini P, et al. Human infections with Plasmodium knowlesi, the Philippines. Emerg Infect Dis. 2008;14:811–3.
Ng OT, Ooi EE, Lee CC, Lee PJ, Ng LC, Pei SW, et al. Naturally acquired human Plasmodium knowlesi infection. Singapore. Emerg Infect Dis. 2008;14:814–6.
Jiang N, Chang Q, Sun X, Lu H, Yin J, Zhang Z, et al. Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010;16:1476–8.
Lubis IN, Wijaya H, Lubis M, Lubis CP, Divis PC, Beshir KB, et al. Contribution of Plasmodium knowlesi to multi-species human malaria infections in North Sumatera. Indonesia. J Infect Dis. 2017;215:1148–55.
Figtree M, Lee R, Bain L, Kennedy T, Mackertich S, Urban M, et al. Plasmodium knowlesi in human, Indonesian Borneo. Emerg Infect Dis. 2010;16:672–4.
Marchand RP, Culleton R, Maeno Y, Quang NT, Nakazawa S. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes, southern Vietnam. Emerg Infect Dis. 2011;17:1232–9.
Iwagami M, Nakatsu M, Khattignavong P, Soundala P, Lorphachan L, Keomalaphet S, et al. First case of human infection with Plasmodium knowlesi in Laos. PLoS Negl Trop Dis. 2018;12:e0006244.
Khim N, Siv S, Kim S, Mueller T, Fleischmann E, Singh B, et al. Plasmodium knowlesi infection in humans, Cambodia, 2007–2010. Emerg Infect Dis. 2011;17:1900–2.
Tyagi RK, Das MK, Singh SS, Sharma YD. Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J Antimicrob Chemother. 2013;68:1081–8.
Hussin N, Lim YAL, Goh PP, William T, Jelip J, Mudin RN. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar J. 2020;19:55.
Cox-Singh J. Zoonotic malaria: Plasmodium knowlesi, an emerging pathogen. Curr Opin Infect Dis. 2012;25:530–6.
Ahmed MA, Cox-Singh J. Plasmodium knowlesi - an emerging pathogen. ISBT Sci Ser. 2015;10:134–40.
Zaw MT, Lin Z. Human Plasmodium knowlesi infections in South-East Asian countries. J Microbiol Immunol Infect. 2019;52:679–84.
Hii J, Vythilingam I, Roca-Feltrer A. Human and simian malaria in the Greater Mekong Subregion and challenges for elimination. In: Manguin S, Dev S, editors. Towards Malaria Elimination - A Leap Forward. Rijeka: InTech; 2018. p. 95–127.
McArthur J. The importance of Anopheles leucosphyrus. Trans R Soc Trop Med Hyg. 1951;44:683–94.
De Zulueta J, Lachance F. A malaria-control experiment in the interior of Borneo. Bull World Health Organ. 1956;15:673–93.
Chang MS, Doraisingam P, Hardin S, Nagum N. Malaria and filariasis transmission in a village/forest setting in Baram District, Sarawak, Malaysia. J Trop Med Hyg. 1995;98:192–8.
Chang MS, Hii J, Buttner P, Mansoor F. Changes in abundance and behaviour of vector mosquitoes induced by land use during the development of an oil palm plantation in Sarawak. Trans R Soc Trop Med Hyg. 1997;91:382–6.
Wharton RH, Eyles DE. Anopheles hackeri, a vector of Plasmodium knowlesi in Malaya. Science. 1961;134:279–80.
Chang MS, Matusop A, Sen FK. Differences in Anopheles composition and malaria transmission in the village settlements and cultivated farming zone in Sarawak, Malaysia. Southeast Asian J Trop Med Public Health. 1999;30:454–9.
Vythilingam I, Tan CH, Asmad M, Chan ST, Lee KS, Singh B. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans R Soc Trop Med Hyg. 2006;100:1087–8.
Jiram AI, Vythilingam I, NoorAzian YM, Yusof YM, Azahari AH, Fong MY. Entomologic investigation of Plasmodium knowlesi vectors in Kuala Lipis, Pahang, Malaysia. Malar J. 2012;11:213.
Wong ML, Chua TH, Leong CS, Khaw LT, Fornace K, Wan-Sulaiman WY, et al. Seasonal and spatial dynamics of the primary vector of Plasmodium knowlesi within a major transmission focus in Sabah, Malaysia. PLoS Negl Trop Dis. 2015;9:10.
Vythilingam I, Lim YA, Venugopalan B, Ngui R, Leong C, Wong M, et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): epidemiologic and entomologic analysis. Parasit Vectors. 2014;7:436.
Sungvornyothin S, Garros C, Chareonviriyaphap T, Manguin S. How reliable is the humeral pale spot for identification of cryptic species of the Minimus Complex? J Am Mosq Control Assoc. 2006;22:185–91.
Paredes-Esquivel C, Donnelly MJ, Harbach RE, Townson H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris subgroup reveals cryptic species: implications for identification of disease vectors. Mol Phylogenet Evol. 2009;50:141–51.
Manguin S, Garros C, Dusfour I, Harbach RE, Coosemans M. Bionomics, taxonomy, and distribution of the major malaria vector taxa of Anopheles subgenus Cellia in Southeast Asia: an updated review. Infect Genet Evol. 2008;8:489–503.
Takano KT, Nguyen NTH, Nguyen BTH, Sunahara T, Yasunami M, Nguyen MD, et al. Partial mitochondrial DNA sequences suggest the existence of a cryptic species within the Leucosphyrus group of the genus Anopheles (Diptera: Culicidae), forest malaria vectors, in northern Vietnam. Parasit Vectors. 2010;3:41.
Sallum MAM, Peyton EL, Harrison BA, Wilkerson RC. Revision of the Leucosphyrus group of Anopheles (Cellia). Rev Bras Entomol. 2005;49:1–152.
Walton C, Handley JM, Kuvangkadilok C, Collins FH, Harbach RE, Baimai V, et al. Identification of five species of the Anopheles dirus complex from Thailand, using allele-specific polymerase chain reaction. Med Vet Entomol. 1999;13:24–32.
Sallum MAM, Foster PG, Li C, Sithiprasasna R, Wilkerson RC. Phylogeny of the Leucosphyrus Group of Anopheles (Cellia) (Diptera: Culcidae) based on mitochondrial gene sequences. Ann Entomol Soc Am. 2007;100:27–35.
Phuc HK, Ball AJ, Son L, Hanh NV, Tu ND, Lien NG, et al. Multiplex PCR assay for malaria vector Anopheles minimus and four related species in the Myzomyia Series from Southeast Asia. Med Vet Entomol. 2003;17:423–8.
Surendran SN, Sarma DK, Jude PJ, Kemppainen P, Kanthakumaran N, Gajapathy K, et al. Molecular characterization and identification of members of the Anopheles subpictus complex in Sri Lanka. Malar J. 2013;12:304.
Goswami G, Singh OP, Nanda N, Raghavendra K, Gakhar SK, Subbarao SK. Identification of all members of the Anopheles culicifacies complex using allele-specific polymerase chain reaction assays. Am J Trop Med Hyg. 2006;75:454–60.
Dusfour I, Blondeau J, Harbach RE, Vythilingham I, Baimai V, Trung HD, et al. Polymerase chain reaction identification of three members of the Anopheles sundaicus (Diptera: Culicidae) complex, malaria vectors in Southeast Asia. J Med Entomol. 2007;44:723–31.
Ma Y, Li S, Xu J. Molecular identification and phylogeny of the Maculatus group of Anopheles mosquitoes (Diptera: Culicidae) based on nuclear and mitochondrial DNA sequences. Acta Trop. 2006;99:272–80.
WHO. WHO Malaria Terminology. Geneva: World Health Organization; 2018.
Rattanarithikul R, Harrison BA, Harbach RE, Panthusiri P, Coleman RE, Panthusiri P. Illustrated keys to the mosquitoes of Thailand. IV. Anopheles. Southeast Asian J Trop Med Public Health. 2006;37(Suppl. 2):1–128.
Reid JA. Anopheline mosquitoes of Malaya and Borneo. Kettering: Staples Printers Limited; 1968.
Rattanarithikul R, Harrison BA, Panthusiri P, Coleman RE. Illustrated keys to the mosquitoes of Thailand I. Background; geographic distribution; lists of genera, subgenera, and species; and a key to the genera. Southeast Asian J Trop Med Public Health. 2005;36:59–69.
Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus-and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687.
Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80.
Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, Wilkerson RC. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst Entomol. 2002;27:361–82.
Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2:103–24.
Sum JS, Lee WC, Amir A, Braima KA, Jeffery J, Abdul-Aziz NM, et al. Phylogenetic study of six species of Anopheles mosquitoes in Peninsular Malaysia based on inter-transcribed spacer region 2 (ITS2) of ribosomal DNA. Parasit Vectors. 2014;7:309.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 2003;52:696–704.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Tan CH, Vythilingam I, Matusop A, Chan ST, Singh B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J. 2008;7:52.
Schultz GW. Biting activity of mosquitos (Diptera: Culicidae) at a malarious site in Palawan, Republic of The Philippines. Southeast Asian J Trop Med Public Health. 1992;23:464–9.
Chua TH, Manin BO, Daim S, Vythilingam I, Drakeley C. Phylogenetic analysis of simian Plasmodium spp. infecting Anopheles balabacensis Baisas in Sabah. Malaysia. PLoS Negl Trop Dis. 2017;11:10.
Hawkes FM, Manin BO, Cooper A, Daim S, Homathevi R, Jelip J, et al. Vector compositions change across forested to deforested ecotones in emerging areas of zoonotic malaria transmission in Malaysia. Sci Rep. 2019;9:13312.
Beckmann JF, Fallon AM. Decapitation improves detection of Wolbachia pipientis (Rickettsiales: Anaplasmataceae) in Culex pipiens (Diptera: Culicidae) mosquitoes by the polymerase chain reaction. J Med Entomol. 2012;49:1103–8.
Nada Raja T, Hu TH, Zainudin R, Lee KS, Perkins SL, Singh B. Malaria parasites of long-tailed macaques in Sarawak, Malaysian Borneo: a novel species and demographic and evolutionary histories. BMC Evol Biol. 2018;18:49.
Gamalo LE, Dimalibot J, Kadir KA, Singh B, Paller VG. Plasmodium knowlesi and other malaria parasites in long-tailed macaques from the Philippines. Malar J. 2019;18:147.
Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 2014;13:68.
Lalremruata A, Magris M, Vivas-Martinez S, Koehler M, Esen M, Kempaiah P, et al. Natural infection of Plasmodium brasilianum in humans: man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine. 2015;2:1186–92.
Imwong M, Madmanee W, Suwannasin K, Kunasol C, Peto TJ, Tripura R, et al. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J Infect Dis. 2019;219:695–702.
Jeslyn WPS, Huat TC, Vernon L, Irene LMZ, Sung LK, Jarrod LP, et al. Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector-Borne Zoonotic Dis. 2011;11:131–5.
Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias. Atlanta: CDC; 2003.
Vidhya PT, Pulikkottil I, Maile A, Zahid AK. Anopheles sundaicus mosquitoes as vector for Plasmodium knowlesi, Andaman and Nicobar Islands, India. Emerg Infect Dis. 2019;25:817–20.
Feachem RGA, Chen I, Akbari O, Bertozzi-Villa A, Bhatt S, Binka F, et al. Malaria eradication within a generation: ambitious, achievable, and necessary. Lancet. 2019;394:1056–112.
Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. Elife. 2014;3:e03401.
Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522.
Google Earth. Map of Lawas, Sarawak. 2018.
Acknowledgements
We would like to thank staff from the Sarawak State Health Department and Lawas District Health Office for their assistance during the field trips. We also thank the Director General of Health Malaysia for permission to publish this article.
Funding
This study was supported by research grants from the Ministry of Higher Education in Malaysia (FRGS/1/2014/SKK01/UNIMAS/03/1), Universiti Malaysia Sarawak (grant no. F05/SpFRC/1433/16/1 and F05/DPP/1508/2016), and by a postgraduate scholarship to JAXD from the Ministry of Higher Education in Malaysia.
Author information
Authors and Affiliations
Contributions
BS designed the study and supervised JXDA, KAK and DSAM while they carried out the molecular studies. BS, PCSD and JXDA analysed the results. KY organised the fieldwork logistics, AM organised and supervised staff from Sarawak Department of Health involved in the field trips and JXDA undertook the fieldwork. BS and JXDA wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of Universiti Malaysia Sarawak and by the Medical Research and Ethics Committee, Ministry of Health Malaysia (NMRR-10-1194-7854). All field staff and volunteers who carried out mosquito collections were provided with antimalarial prophylaxis.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Plasmodium SSU rDNA sequences and their GenBank accession numbers.
Additional file 2: Table S2.
cox1 and ITS2 sequences of Anopheles mosquitoes and their GenBank accession numbers.
Additional file 3: Alignment S1.
Multiple sequence alignment of the cox1 gene of An. barbirostris (s.l.) and an outgroup, An. coustani.
Additional file 4: Alignment S2.
Multiple sequence alignment of the cox1 gene of An. leucosphyrus (s.l.) and an outgroup, An. gambiae.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ang, J.X.D., Kadir, K.A., Mohamad, D.S.A. et al. New vectors in northern Sarawak, Malaysian Borneo, for the zoonotic malaria parasite, Plasmodium knowlesi. Parasites Vectors 13, 472 (2020). https://doi.org/10.1186/s13071-020-04345-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04345-2