Abstract
Background
Almost 1000 cases of American cutaneous leishmaniasis have been registered yearly in Rondônia State, Brazil. Little is known about the Leishmania transmission cycle (vectors and reservoirs) in the state. This study aimed to evaluate sand fly fauna from two vertical stratification layers in order to identify potential vectors and their blood-meal sources.
Methods
The study was conducted in Jamari National Forest. Sand flies were collected in the canopy (15 m) and at ground level (1 m) using HP light traps during four months, February, April, August and October, 2018. Insects were identified to the species level, and females were subjected to DNA extraction and PCR targeting minicircle kDNA and hsp70 (for Leishmania detection and species identification), and cytb (to identify blood-meal sources). Exploratory data analysis was used to determine mean of abundance and species richness between stratifications. The hsp70 and cytb sequences were analyzed and compared with sequences from GenBank.
Results
Overall, 68 species were identified from 15,457 individuals. On the Potosi trail, 7531 individuals of 49 species were collected; canopy captures totaled 6463 individuals of 46 species, while ground captures totaled 1068 individuals of 38 species. On the Santa Maria trail, 7926 individuals of 61 species were collected; canopy captures totaled 6136 individuals of 51 species, while ground captures totaled 1790 individuals of 53 species. A total of 23 pools were positive for kDNA (canopy n = 21, ground n = 2). Only two samples were sequenced for hsp70 (both in canopy); one sequence exhibited similarity with Leishmania braziliensis (Lutzomyia davisi pool) and another with L. naiffi (Lu. antunesi pool). The cytb fragment was amplified in 11 of 86 samples. Sample sequencing identified cytb DNA from 5 blood-meal sources: Micrastur gilvicollis, Psophia viridis, Tamandua tetradactyla, Homo sapiens and Choloepus didactylus.
Conclusions
Sand fly fauna is more diverse in the canopy than at ground level. Factors such as blood-meal sources, resting sites, and abiotic components probably contribute to high abundance in the canopy. Our results reinforce the possibility that Lu. antunesi and Lu. davisi participate in Leishmania transmission in forest environments and may play an important role in transmission from sylvatic to human hosts.

Similar content being viewed by others
Background
Phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae) are small dipteran insects that play a role in the transmission of Leishmania species, the protozoans causing leishmaniases [1]. There are approximately 1000 phlebotomine sand fly species in the world; 286 occur in Brazil and 13 of these act as vectors [2].
Cutaneous leishmaniasis (CL) is characterized by localized lesions (LCL), but may also take the form of mucocutaneous leishmaniasis (ML), disseminated cutaneous leishmaniasis (DCL) or anergic diffuse cutaneous leishmaniasis (ADCL) [3]. This disease has a significant impact on public health; it is endemic in 98 countries with 0.7–1.2 million people infected per year and an estimated 350 million people at risk of contracting the disease [4]. In 2018 alone, Brazil recorded approximately 17,000 cases of Leishmaniasis caused by seven Leishmania species: Leishmania amazonensis; L. braziliensis (the most prevalent); L. guyanensis; L. lainsoni; L. lindenbergi; L. naiffi; and L. shawi [5, 6].
In 2018, 1018 cases of CL were recorded in the state of Rondônia, Brazil [6]. CL has a zoonotic transmission cycle in which sand flies transmit Leishmania parasites between wild vertebrates. Humans are considered accidental hosts and most CL cases in Rondônia have been associated with anthropic activities (such as hunting, fishing, logging and mining) which are conducted in or near forest environments [7, 8]. Human cases of CL are caused by seven Leishmania species [8,9,10], and sand flies have been found carrying three of these species: L. amazonensis, L. braziliensis and L. naiffi [11,12,13].
To understand how CL is transmitted in the region, it is necessary to determine ecological parameters such as the composition and diversity of phlebotomine species (with a focus on vectors) and to identify which vertebrate blood meals participate in the maintenance of the Leishmania cycle in forest environments. In rainforests, the distribution of sand fly populations can be influenced by physical, biological and microclimatic conditions that differ between stratification levels [14]. For example, Lutzomyia davisi is found in abundance both in canopy and at ground level [14, 15], but Lutzomyia flaviscutellata tends to be distributed at ground level where it feeds primarily on rodents [16, 17].
Since 2016, our study group has been conducting surveys to assess vector species diversity and the prevalence of Leishmania parasites in Rondônia [12, 13, 18, 19]. To date, 143 sand fly species have been recorded in Rondônia [20] and Lu. davisi has been identified as a potential vector because it occurs in high abundance and field-collected females of the species have been tested positive for Leishmania DNA [10, 11, 13]. Few studies conducted in Rondônia have compared sand fly distribution patterns between stratification levels [12, 21] and little is known about Leishmania reservoirs in the region [10]. In a previous study, we used molecular methods to target the cytb region and detected human, bovine and anteater DNA in sand flies collected from three different environments [13].
This study continues that line of research by describing the composition and distribution of sand fly species in two different stratifications (canopy and ground), and by aiming to: assess relative richness and abundance, detect natural infection with Leishmania DNA, and identify sand fly blood-meal sources in the natural environment.
Methods
Study area
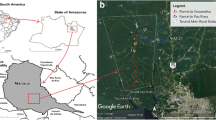
Rondônia State is in the North Region of Brazil (Fig. 1a); it has an area of 237,576.2 km2, 52 municipalities and a population of 1,768,204. It has an equatorial dry and wet climate which generally entails a dry season between June and August and a rainy season between October and April, while May and September are months of seasonal transition. The average annual precipitation is 2000 mm; the relative humidity is 80–85%, and the average temperature 24 °C [22].
This study was performed in Jamari National Forest (Jamari Flona), located in northern Rondônia State, in the municipality of Itapuã do Oeste (Fig. 1b, c). This forest became a conservation unit on September 25, 1984. Jamari Flona has an area of approximately 225,000 ha [23]. The phytophysiognomy is composed of 90% ombrophilous dense forest [24]. Jamari Flona was a site of ore extraction in the 1950s. Part of this reserve has been designated as a Management Forest Unit (MFU) and part has been set aside for permanent preservation [23].
The Flona contains trails with 5 km in length used by the Chico Mendes Institute of Biodiversity Conservation (ICMBIO) under the National Programme of Biodiversity Knowledge. On the basis of yearly studies, we selected two trails for survey [12]: the Potosi trail (09°15′36.14″S, 62°54′48.33″W) and the Santa Maria trail (09°08′22.65S, 62°54′49.04″W) (Fig. 1d).
Sand fly collection was authorized by ICMBIO under SISBIO system number 58563-3, and by the National System of Genetic Heritage Management and Associated Knowledge (SISGEN) under code AA32B8E, “Studies about Amazonian sand flies.”
Sand fly collection and identification
Sand fly collections were performed for 5 consecutive nights in the months of February, April, August, and October, 2018. HP® light traps [25] (Biomedica, Belo Horizonte, Brazil) were installed (from 18:00 to 7:00 h) at 8 points on the Santa Maria trail. The first point was positioned 500 m from the trailhead and the second point was positioned 300 m from the first point, and the remaining points were equally distanced from each other (at 300 m intervals) (Fig. 1d). At each point, traps were installed at ground level (1 m above ground) and in the canopy (15 m above ground); 16 HP traps were installed in total. In the canopy, traps were suspended from a “slingshot”, i.e. a fishing line (0.40 mm) weighted with a lead ball (20 g) and secured with a nylon rope (4 mm). The same design was replicated simultaneously on the Potosi trail. Mesh was not used in the HP light traps; instead, traps were outfitted with BD FalconTM 50 ml conical centrifuge tubes (BD Company, Bedford, USA) containing 96% ethanol.
After each night of collection, sand flies were separated from other insects using aspirators and, after chilling, examined under a binocular microscope at 5–20× magnification. Thereafter, the sand flies were stored in 96% ethanol, and maintained at a low temperature in loci. After each month of collection, specimens were placed in boxes containing ice in order to maintain a low temperature during transport to the Entomology Laboratory of Fiocruz Rondônia, Porto Velho. Males were mounted in Berleseʼs medium [26]. The head and last 3 segments of each female were mounted, and the rest of the body was stored in 96% ethanol until molecular processing. Species identification was performed using the taxonomic keys by Young & Duncan [27], and Galati [28]. We adopted the nomenclatural of Young & Duncan [27] and the generic abbreviations proposed by Marcondes [29].
Molecular detection of Leishmania
Non-engorged females of the most abundant species were sorted according to trail and stratification layer and separated into pools of 2–20 specimens. Pools were subjected to DNA extraction and PCR assays targeting minicircle kDNA using the primers 5′-GGG (G/T)AG GGG CGT TCT (G/C)CG AA-3′ and 5′-(G/C)(G/C)(G/C) (A/T)CT AT(A/T) TTA CAC CAA CCC C-3′ (120 bp). Positive samples from the first reaction were subjected to PCR targeting the hsp70 region using the primers 5′-GGA CGA GAT CGA GCG CAT GGT-3′ and 5′-TCC TTC GAC GCC TCC TGG TTG-3′ (240 bp), as described elsewhere [12, 30]. Lutzomyia ubiquitalis males and the L. amazonensis reference strain IOC/L0575 (IFLA/BR/1967/PH8) were used as the positive controls and ultrapure water was used as the negative control.
Blood-meal identification
Engorged females were separated according to species, trail and strata. Three samples were used as the negative control during DNA extraction:1 sample containing DNA-free water and 2 samples containing a female sand fly with no blood present in the gut. DNA extraction followed the phenol/chloroform protocol described by Sambrook & Russell [31]. PCR used the primers cytb 1 and cytb 2, which are complementary to the conserved region of the cytochrome b gene in vertebrate mtDNA [32].
The amplification reactions (total volume of 50 µl) contained 25 µl (1×) Go Taq Colorless (Promega, Madison, USA), 1.5 µl of each primer (cytb 1 and cytb 2, 10 µM) and 5 µl of DNA (< 250 ng). Amplifications were performed in a thermocycler (Veriti®; Applied Biosystems, Foster City, USA) with an initial denaturation of 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 53 °C for 30 s and extension at 72 °C for 1 min, with a final extension step at 72 °C for 6 min. Amplified products were purified using the QIAquick Purification Kit (Qiagen, Hilden, Germany) and submitted to the Fiocruz Sequencing Facility (RTP01E, Belo Horizonte, Brazil)
Data analysis
Exploratory data analysis (EDA) was used to describe the abundance and total mean richness of the species collected between strata (canopy × ground). Mean abundance and mean species richness were computed per trap per night. Minimum infection rates were obtained by multiplying the number of kDNA positive pools by 100 and dividing the result by the total number of females in these pools [12]. The proportion of engorged to non-engorged females was determined for the most abundant species from each stratification. All data were analyzed using the R statistical environment [33] (R scripts are described in Additional file 1: Dataset S1, Additional file 2: Dataset S2).
The hsp70 and cytb genes were sequenced in duplicate for each sample and the sequences obtained were assembled and analyzed using the Staden package [34] based on Phred values of 30 or above [35]. The consensus sequences were submitted to a BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared with sequences from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). All new sequences generated in this study were deposited in the GenBank database under the accession numbers MT234139, MT260076-MT7260081, MT293514-MT7293518 and MT300189.
Results
A total of 15,457 individuals comprising 68 species were collected. Some females were identified only at the subgenus level: Lutzomyia (Trichophoromyia) sp. (n = 30) and Lutzomyia (Trichopygomyia) sp. (n = 339). The most abundant species were Lutzomyia antunesi (n = 2530), Lu. ayrozai (n = 2198), Lu. davisi (n = 2019), Lu. yuilli yuilli (n = 1483) and Lu. ubiquitalis (n = 1153) (Table 1). The sex ratio of the potential vectors is outlined in Table 2.
Diversity between stratifications
On the Potosi trail, 7531 individuals were collected, and 49 species were identified. Eleven species were recorded solely in canopy, 3 species were recorded solely at ground level, and 34 species were collected in both strata (Table 1).
In the canopy 6463 individuals were collected, and 46 species were identified. In the canopy, the highest mean abundance per trap/night occurred in August (32.1 individuals), followed by February (31.3), October (18.6) and April (10.4). The highest mean richness captured per trap/night occurred in February (5.6 species), followed by October (4.9), April (4.1) and August (2.9) (Fig. 2a). The most abundant species in the canopy were Lu. antunesi (1873), Lu. ayrozai (1256), Lu. y. yuilli (848) and Lu. davisi (521) (Table 1).
At the ground level, 1068 individuals were collected, and 38 species were identified. The highest mean abundance per trap/night occurred in October (8.6 individuals), followed by February (6.9), August (3.7) and April (3.3). The highest mean richness captured per trap/night occurred in October (3.6 species), followed by February (3.4), April (2.3) and August (2.0) (Fig. 2a). The most abundant species at ground level were Lu. tarapacaensis (171), Lu. ubiquitalis (132), Lu. davisi (121) and Lu. ayrozai (110).
On the Santa Maria trail, 7926 individuals were collected, and 61 species were identified. Eight species were recorded solely in canopy, 10 species were recorded solely at ground level, and 43 species were collected in both strata (Table 1).
In the canopy, 6136 individuals were collected, and 51 species were identified. The highest mean abundance per trap/night occurred in February (40.2 individuals), followed by August (23.4), April (14.3) and October (11.5). The highest mean richness captured per trap/night occurred in February (6.7 species), followed by October (5.2), April (5.1) and August (3.9) (Fig. 2b). The most abundant species in the canopy were Lu. davisi (1225), Lu. ayrozai (786), Lu. furcata (764), Lu. y. yuilli (531), and Lu. antunesi (530) (Table 1).
At the ground level, 1790 individuals were collected, and 53 species were identified. The highest mean abundance occurred in February (11.1 individuals), followed by April (9.3), October (8.4) and August (4.9). The highest mean richness occurred in October (4.7 species), followed by August (3.4), February (3.2) and April (2.5) (Fig. 2b). The most abundant species at ground level was Lu. ubiquitalis (780).
Detection of Leishmania DNA
A total of 2817 sand fly females were sorted into 194 pools (Table 2): 67 pools from Santa Maria trail (canopy: 52 pools; ground: 15 pools) and 127 pools from Potosi trail (canopy: 105 pools; ground: 22 pools). The 120-bp minicircle kDNA fragment was amplified in 23 pools: 6 from Santa Maria trail (all from canopy) and 17 from Potosi trail (canopy: 15; ground: 2). The minimal infection rate was 0.81% (23/2817). The 240 bp hsp70 fragment was amplified in 8 pools: 5 pools from the Santa Maria trial and 3 pools from the Potosi trail. Sequencing was successful for 2 samples in which the hsp70 fragment was amplified. Both samples were from the canopy level of the Santa Maria trail; 1 sequence exhibited similarity with L. braziliensis (Lu. davisi pool) and the other exhibited similarity with L. naiffi (Lu. antunesi pool) (Table 3).
Blood-meal identification
Of the 8788 females collected 86 (0.97%) were engorged: 28 from the Santa Maria trail (canopy: 22; ground: 6) and 58 from the Potosi trail (canopy: 45; ground: 13). The most abundant species with blood present in the gut were Lu. ayrozai (n = 24), Lu. antunesi (n = 7) and Lu. davisi (n = 6) from canopy (Fig. 3a–c) and Lu. davisi (n = 3), Lu. tarapacaensis (n = 3), Lu. ayrozai (n = 2), Lu. antunesi (n = 2) and Lu. y. yuilli (n = 2) from the ground level (Fig. 3b–d).
Amplification of the cytb region was attempted for all 86 samples and successful identification was achieved for 11 individuals (0.12%): Lu. antunesi (n = 4); Lutzomyia fiocruzi (n = 2); Lu. ayrozai (n = 2); and Lu. davisi (n = 3). The sequenced amplification products exhibited similarity with the cytb DNA of anteaters (Tamandua tetradactyla), humans (Homo sapiens), sloths (Choloepus didactylus) and two species of bird: the lined forest falcon (Micrastur gilvicollis); and the green-winged trumpeter (Psophia viridis) (Table 4).
Discussion
We evaluated the abundance and richness patterns of sand fly fauna in two stratifications in Jamari Flona, Rondônia, Brazil. Abundance was highest in the canopy, where approximately 60% of individuals belonged to four species, Lu. antunesi, Lu. y. yuilli, Lu. ayrozai and Lu. davisi. These findings corroborate those of Resadore et al. [12] who surveyed the same locality and found Lu. y. yuilli and Lu. davisi to be the most abundant species in the canopy; however, at ground level we found Lu. ubiquitalis to be the most abundant species while Resadore et al. [12] found this species in low abundance in both strata. Souza et al. [36] also found Lu. ubiquitalis in abundance at ground level, which may indicate that the distribution of this species is specific to ground level habitats.
For each month of collection on both trails, the mean abundance and richness of sand flies per trap/night was higher in the canopy than at ground level. This finding corroborates the findings of other Amazonian studies [12, 14, 16]. The canopy provides more plentiful resting sites among leaves and trunks, and offers a greater variety of nocturnal blood-meal sources which may attract sand flies since sand flies are most active at night. Both factors, blood meals and resting sites, may contribute to the presence of sand flies in the upper stratum [17].
Overall, abundance was highest in February and October, which is typical since sand flies tend to be captured mostly during the rainy season [17, 36, 37]. In Rondônia, the rainy season generally begins in October and peaks between January and April [22]. In the Central Amazon, between 1977 and 1978, Arias & Freitas [17] observed the highest number of sand fly captures in November, December and May, but these authors speculated that the high number of captures may have resulted from a high rate of adult emergence among Lu. anduzei and Lu. umbratilis.
We were surprised to find that sand fly abundance was higher in August than in October and April in the canopy of the Potosi trail, even though August is part of the dry season. Godoy et al. [37] verified a negative correlation between precipitation and the abundance of the Lu. antunesi in the municipality of Guaraí, Tocantins State; thereby demonstrating that precipitation can influence the abundance of sand fly populations. Our study did not account for abiotic variables and our sample design did not allow us to evaluate the impact of seasonality; therefore, it was difficult to attribute a cause to the high level of sand fly abundance observed in the dry month of August. Further studies will need to determine which factors influenced the capture rates that we observed.
The presence of Leishmania species and the identity of blood-meal sources were determined for sand flies from both strata. For Leishmania detection, of the 23 pools that were positive for minicircle kDNA and the eight pools in which hsp70 was amplified, only two samples were successfully identified by sequencing. The kDNA region has a high number of copies and is more sensitive to amplification, but it is present in the mitochondrial DNA of the family Trypanosomatidae and thus does not permit specific identification of Leishmania; the hsp70 region does permit specific identification of Leishmania species, but it has fewer copies and is less sensitive to amplification [30]. In this study, the identification of six hsp70-positive samples was not possible due to the low quality of the sequences.
The Leishmania species identified here are L. braziliensis and L. naiffi. Currently in Rondônia, L. braziliensis is the most prevalent species of Leishmania associated with human cases of leishmaniasis and it has been found previously in sand flies [10, 11, 13]. Leishmania braziliensis has a variety of hosts [38]; thus, different sand fly species with different blood-meal sources could nevertheless acquire the same parasite from a variety of vertebrate hosts, which may account for the presence of L. braziliensis in many sand fly species. We found L. braziliensis DNA in Lu. davisi, which corroborates the findings of other studies conducted in Rondônia [10, 13] and reinforces the possibility that Lu. davisi is a putative vector of L. braziliensis.
Leishmania naiffi is uncommon in human cases but some studies indicate the possibility that L. naiffi infection has been underreported in the North Region of Brazil [39]. The natural host of L. naiffi is Dasypus novemcinctus [40]. In humans, infection with L. naiffi generally manifests as localized lesions that are amenable to spontaneous cure or rapid treatment [39, 41,42,43]. Leishmania naiffi has been identified in only one human case of leishmaniasis in Rondônia, a 35 year-old man from the municipality of Rolim de Moura [43].
In Rondônia, L. naiffi has been found in field-collected females of the Lu. ayrozai [5], also females of Lu. davisi and Lu. hirsuta hirsuta have been found carrying L. naiffi flagellates [10, 11]. In this study, no Lu. ayrozai females were found with Leishmania DNA, but this is the first time that L. naiffi has been identified in Lu. (Nyssomyia) sand flies from Rondônia. Lutzomyia antunesi is abundant in many parts of the state and may act as a vector there [11, 13, 19]; however, to date, Lu. antunesi is a proven vector only of L. lindenbergi in Pará State [44]. Given that Lu. davisi and Lu. antunesi are frequently found in abundance in Rondônia, further studies need to examine colonization patterns and the vector competence and capacity of these species in relation to Leishmania transmission cycles.
Of the 8788 females collected, 86 were engorged (less than 1% of the total). This proportion is in line with two previous studies, one in which 15 out of 4089 females were engorged [13], and another in which seven out of 708 females were engorged [45]. These extremely small proportions suggest that light traps are not an effective method for collecting engorged females.
Blood-meal identification was possible for only 0.12% of total females captured. This decrease (from 1% to 0.12%) may be due to differing stages of digestion among engorged females. After blood-feeding, the blood meal is generally directed to the midgut where enzymatic activity increases, and this step may degrade the DNA and thus reduce PCR sensitivity [46]. For example, in one study the efficacy of amplification was reduced by 20% in mosquito samples that were submitted to PCR (targeting the cytb gene) 33 h after blood-feeding [47], and, in another study, Baum et al. [48] subjected blood from 93 engorged females to PCR targeting the PNOC partial gene 24 h after blood-feeding, and blood-meal identification was possible in only 27 females. These observations demonstrate that more effective technologies need to be developed for the detection of blood-meal sources.
Our findings improve the knowledge of sand fly blood-meal sources in sylvatic environments. The abundance of Lu. antunesi, Lu. ayrozai and Lu. davisi in the canopy may indicate that these species blood-feed on arboreal animals. In a study conducted in the Central Amazon, Arias et al. [17] found that when a natural host is ground-dwelling, the associated sand fly species feed at ground level, and when the natural host is canopy-dwelling the associated species feed at canopy level. However, given the low sample size obtained in our collections and given that anteaters, sloths, and birds frequently move between canopy and ground level, it was not possible to determine whether or not feeding was occurring arboreally. Similarly, although human DNA was found in a Lu. davisi female collected in the canopy, it was impossible to determine if this species favors blood meals from a specific stratum. We believe that this female fed on the ground and was later captured in the canopy.
The data presented here may improve local knowledge of ACL epidemiology. This is significant because much of the local populous works in the Amazonian forests [6, 8] where the sylvatic transmission cycle occurs, and therefore these people experience greater exposure to Leishmania-infected vectors.
Conclusions
Our study demonstrates that sand fly abundance and richness is higher in the canopy than at ground level, and these findings corroborate other vertical stratification studies. Certain factors, including the presence of blood meals, most likely influence sand fly distribution between strata. Our findings also corroborate earlier studies conducted in Rondônia State which indicate that Lu. antunesi and Lu. davisi play a role as vectors [10, 11, 13, 18, 19]. The fact that these species are present in both strata may indicate that they contribute to parasite transmission between strata. This information augments our knowledge of sand flies in Rondônia and may help improve leishmaniasis surveillance and control programmes.
Availability of data and materials
Data supporting the conclusions of this article are included within the article and its additional files. The newly generated sequences were deposited in the GenBank database under the accession numbers MT234139, MT260076-MT7260081, MT293514-MT7293518 and MT300189. Raw data are available without restriction upon request.
Abbreviations
- CL:
-
cutaneous leishmaniasis
- LCL:
-
localized lesions (LCL)
- ML:
-
mucocutaneous leishmaniasis
- DCL:
-
disseminated cutaneous leishmaniasis
- ADCL:
-
anergic diffuse cutaneous leishmaniasis
- cytb :
-
cytochrome b
- kDNA:
-
kinetoplast DNA
- hsp70:
-
heat-shock protein 70
- MFU:
-
Management Forest Unit
- ICMBIO:
-
Instituto Chico Mendes Institute for Biodiversity Conservation
- SISBIO:
-
Biodiversity Authorization and Information System
- SISGEN:
-
National System of Genetic Heritage Management and Associated Knowledge
- PCR:
-
polymerase chain reaction
- mtDNA:
-
mitochondrial DNA
- EDA:
-
exploratory data analysis
References
Ready PD. Biology of phlebotomine sand flies as vectors of disease agents. Annu Rev Entomol. 2013;58:227–50.
Rangel EF, Lainson R, Carvalho BM, Costa SM, Shaw JJ. Sand fly vectors of American cutaneous leishmaniasis in Brazil. In: Rangel EF, Shaw JJ, editors. Brazilian sand flies: biology, taxonomy, medical importance and control. Gewerbestrasse: Springer; 2018. p. 341–80.
Scorza B, Carvalho E, Wilson M. Cutaneous manifestations of human and murine leishmaniasis. Int J Mol Sci. 2017;18:1296.
Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671.
Lainson R. The neotropical Leishmania species: a brief historical review of their discovery, ecology and taxonomy. Rev Pan-Amazônica Saúde. 2010;1:13–32.
Datasus. Sistema de informações de agravos de notificação. Epidemiológicas e Morbidade. Leishmaniose Tegumentar Americana. Portal Ministério da Saúde; 2018. http://www2.datasus.gov.br/DATASUS/index.php?area=0203&id=29892200&VObj=http://tabnet.datasus.gov.br/cgi/deftohtm.exe?sinannet/cnv/lta. Accessed 20 Jan 2018.
Confalonieri UEC, Margonari C, Quintão AF. Environmental change and the dynamics of parasitic diseases in the Amazon. Acta Trop. 2014;129:33–41.
Shaw JJ, Faria DL, Basano SA, Corbett CE, Rodrigues CJ, Ishikawa EEA, et al. The aetiological agents of American cutaneous leishmaniasis in the municipality of Monte Negro, Rondônia state, western Amazonia, Brazil. Ann Trop Med Parasitol. 2007;101:681–8.
Cantanhêde LM, da Silva Júnior CF, Ito MM, Felipin KP, Nicolete R, Salcedo JMV, et al. Further evidence of an association between the presence of Leishmania RNA virus 1 and the mucosal manifestations in tegumentary leishmaniasis patients. PLoS Negl Trop Dis. 2015;9:e0004079.
Grimaldi G, Momen H, Naiff RD, McMahon-Pratt D, Barrett TV. Characterization and classification of leishmanial parasites from humans, wild mammals, and sand flies in the Amazon Region of Brazil. Am J Trop Med Hyg. 1991;44:645–61.
Gil LHS, Basano SA, Souza AA, Silva MGS, Barata I, Camargo LMA, et al. Recent observations on the sand fly (Diptera: Psychodidae) fauna of the State of Rondônia, western Amazônia, Brazil: the importance of Psychodopygus davisi as a vector of zoonotic cutaneous leishmaniasis. Mem Inst Oswaldo Cruz. 2003;98:751–5.
Resadore F, Pereira Júnior AM, Paulo PFM, Gil LHS, Rodrigues MMS, Araújo MS, et al. Composition and vertical stratification of phlebotomine sand fly fauna and the molecular detection of Leishmania in forested areas in Rondônia State Municipalities, western Amazon, Brazil. Vector Borne Zoonotic Dis. 2018;19:347–57.
Pereira Júnior AM, Souza ABN, Castro TS, Silva MS, Paulo PFM, Ferreira GEM, et al. Diversity, natural infection and blood-meal sources of phlebotomine sandflies (Diptera, Psychodidae) in the western Brazilian Amazon. Mem Inst Oswaldo Cruz. 2019;114:e190170.
Dias Lima AG, Bermúdez EC, Medeiros JF, Sherlock I. Estratificação vertical da fauna de flebótomos (Diptera, Psychodidae) numa floresta primária de terra firme da Amazônia Central, Estado do Amazonas, Brasil. Cad Saúde Pública. 2002;18:823–32.
Feitosa MAC, Castellón EG. Fauna de flebotomíneos (Diptera: Psychodidae) em fragmentos de floresta ao redor de conjuntos habitacionais na cidade de Manaus, Amazonas, Brasil. I. Estratificação Vertical. Acta Amaz. 2006;36:539–48.
Arias JR, Freitas RA. Flebotomíneos da Amazônia Central do Brasil. 1: Resultados obtidos das capturas feitas com isca humana e eqüina (Diptera: Psychodidae). Acta Amaz. 1977;7:507–27.
Arias JR, Freitas RA. On the vectors of cutaneous leishmaniasis in central amazon of Brasil. 3. Phlebotomine sand fly stratification in a terra firme forest. Acta Amaz. 1982;12:599–608.
Ogawa GM, Pereira Júnior AM, Resadore F, Ferreira RGM, Medeiros JF, Camargo LMA. Sandfly fauna (Diptera: Psychodidae) from caves in the state of Rondônia, Brazil. Rev Bras Parasitol Veterinária. 2016;25:61–8.
Resadore F, Pereira Júnior AM, Carvalho LPC, Dos Santos APA, Teles CBG, Medeiros JF. Phlebotomine sand fly composition (Diptera: Psychodidae) and putative vectors of American cutaneous leishmaniasis in Porto Velho Municipality, western Amazon, Brazil. J Med Entomol. 2017;54:798–803.
Costa GS, Pereira Júnior AM, Pessoa FAC, Shimabukuro PHF, Medeiros JF. New records of phlebotomine sand flies (Diptera: Psychodidae) from the western Brazilian Amazon and the description of the female of Pintomyia fiocruzi. J Med Entomol. 2020;57:1328–33.
Azevedo ACR, Luz SLB, Vilela ML, Rangel EF. Studies on the sandfly fauna of Samuel Ecological Station, Porto Velho Municipality, Rondônia State, Brazil. Mem Inst Oswaldo Cruz. 1993;88:509–12.
Bezerra RB, Dantas RT, Trindade AG. Caracterização temporal da precipitação pluvial do município de Porto Velho/RO no período de 1945 a 2003. Soc Nat. 2010;22:609–23.
Ministério do Meio Ambiente. Floresta Nacional do Jamari - Rondônia. 1st ed. Brasília: Ministério do Meio Ambiente/IBAMA; 2005.
Instituto Brasileiro de Geografia e Estatística. Manual Técnico da Vegetação Brasileira. 2nd ed. Rio de Janeiro: Diretoria de Geociências do IBGE; 2012.
Pugedo H, Barata RA, França-Silva JC, Silva JC, Dias ES. HP: um modelo aprimorado de armadilha luminosa de sucção para a captura de pequenos insetos. Rev Soc Bras Med Trop. 2005;38:70–2.
Langeron M. Précis de Microscopie. 7th ed. Paris: Masson, Libraires de L’Académie de Medicine; 1949.
Young DG, Duncan MA. Guide to the identification and geographic distribution of Lutzomyia sandflies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae). Mem Am Entomol Inst. 1994;54:1–881.
Galati EAB. Phlebotominae (Diptera, Psychodidae): classification, morphology and terminology of adults and identification of American taxa. In: Rangel EF, Shaw JJ, editors. Brazilian sand flies: biology, taxonomy, medical importance and control. Gewerbestrasse: Springer; 2018. p. 9–212.
Marcondes CB. A proposal of generic and subgeneric abbreviations for phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae) of the world. Entomol News. 2007;118:351–6.
Graça GC, Volpini AC, Romero GAS, Oliveira Neto MP, Hueb M, Porrozzi R, et al. Development and validation of PCR-based assays for diagnosis of American cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz. 2012;107:664–74.
Sambrook J, Russell D. Isolament and quantification of DNA. In: Inglis J, Boyle A, Gann A, editors. Molecular cloning, a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2012. p. 1–78.
Boakye DA, Tang J, Truc P, Merriweather A, Unnasch TR. Identification of bloodmeals in haematophagous Diptera by cytochrome B heteroduplex analysis. Med Vet Entomol. 1999;13:282–7.
R Development Core Team. A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. http://www.r-project.org.
Staden R. The staden sequence analysis package. Mol Biotechnol. 1996;5:233–41.
Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94.
Souza AAA, Santos TV, Jennings YLL, Ishikawa EAY, Barata IR, Silva MGS, et al. Natural Leishmania (Viannia) spp. infections in phlebotomine sand flies (Diptera: Psychodidae) from the Brazilian Amazon region reveal new putative transmission cycles of American cutaneous leishmaniasis. Parasite. 2016;23:22.
Godoy RE, Santana ALF, Rangel EF, Graser C, Vilela ML. Aspects on the ecology of Phlebotomine sand flies (Diptera: Psychodidae) from Guaraí, State of Tocantins, Brazil, endemic area for American cutaneous leishmaniasis. J Med Entomol. 2017;54:229–35.
Roque ALR, Jansen AM. Wild and synanthropic reservoirs of Leishmania species in the Americas. Int J Parasitol Parasites Wildl. 2014;3:251–62.
Fagundes-Silva GA, Romero GAS, Cupolillo E, Yamashita EPG, Gomes-Silva A, Guerra JAO, et al. Leishmania (Viannia) naiffi: rare enough to be neglected? Mem Inst Oswaldo Cruz. 2015;110:797–800.
Lainson R, Shaw JJ. Leishmania (Viannia) naiffi sp. n., a parasite of the armadillo, Dasypus novemcinctus (L.) in Amazonian Brazil. Ann Parasitol Hum Comp. 1989;64:3–9.
Naiff RD, Freitas RA, Naiff MF, Arias JR, Barrett TV, Momen H, et al. Epidemiological and nosological aspects of Leishmania naiffi Lainson & Shaw, 1989. Mem Inst Oswaldo Cruz. 1991;86:317–21.
Camara Coelho LI, Paes M, Guerra JAO, Barbosa MG, Coelho C, Lima B, et al. Characterization of Leishmania spp. causing cutaneous leishmaniasis in Manaus, Amazonas, Brazil. Parasitol Res. 2011;108:671–7.
Figueira LP, Soares FV, Júnior RDN, Vinhote-Silva AC, Silva SS, Espir TT, et al. New human case reports of cutaneous leishmaniasis by Leishmania (Viannia) naiffi in the Amazon region, Brazil. Acta Amaz. 2017;47:47–52.
Silveira FT, Ishikawa EAY, De Souza AAA, Lainson R. An outbreak of cutaneous leishmaniasis among soldiers in Belém, Pará State, Brazil, caused by Leishmania (Viannia) lindenbergi n. sp. A new leishmanial parasite of man in the Amazon region. Parasite. 2002;9:43–50.
Torchitte APA, Pereira Júnior AM, Paulo PFM, Costa GS, Castro TS, Ferreira RGM, et al. Identification of sand flies (Diptera: Psychodidae) and blood-meal sources in periurban areas of Ji-Paraná municipality, western Brazilian Amazon. Braz J Biol. 2020. (In press).
Haouas N, Pesson B, Boudabous R, Dedet JP, Babba H, Ravel C. Development of a molecular tool for the identification of Leishmania reservoir hosts by blood meal analysis in the insect vectors. Am J Trop Med Hyg. 2007;77:1054–9.
Oshaghi MA, Chavshin AR, Vatandoost H, Yaaghoobi F, Mohtarami F, Noorjah N. Effects of post-ingestion and physical conditions on PCR amplification of host blood meal DNA in mosquitoes. Exp Parasitol. 2006;112:232–6.
Baum M, Alcântara E, Pinto M, Marchi T, Baura W, Klisiowicz D, et al. Molecular detection of the blood meal source of sand flies (Diptera: Psychodidae) in a transmission area of American cutaneous leishmaniasis, Paraná State, Brazil. Acta Trop. 2014;143:8–12.
Acknowledgements
We would like to thank to members of Jamari FLONA by the support in this study.
Funding
Fundação de Amparo ao Desenvolvimento das Ações Científicas e Tecnológicas e à Pesquisa do Estado de Rondônia (FAPERO) - Chamada 012/2016 - Diversidade Genética de Flebotomíneos no Estado de Rondônia e Avaliação dos Ciclos de Transmissão de Leishmanioses. This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Financial code 001.
Author information
Authors and Affiliations
Contributions
POL and AMPJ contributed equally to this work, performed the sample collection, identified the insects, and wrote the manuscript; ABNS, MSS and TSC participated in the preparation of sand flies for molecular analysis; MMSR developed the sample design and performed the data analysis; MTSF participated in the bioinformatics analysis; LPCC performed the sample collection; PFMP performed the molecular biology assays; GEMF and JFM designed the study and helped draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Dataset S1.
R scripts.
Additional file 2: Dataset S2.
R help functions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Leão, P.d., Pereira Júnior, A.M., de Paulo, P.F.M. et al. Vertical stratification of sand fly diversity in relation to natural infections of Leishmania sp. and blood-meal sources in Jamari National Forest, Rondônia State, Brazil. Parasites Vectors 13, 422 (2020). https://doi.org/10.1186/s13071-020-04295-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04295-9