Abstract
Background
Triatoma sordida, a vector of Trypanosoma cruzi, is native of Brazil, Bolivia, Paraguay, Argentina, and Uruguay, and occurs primarily in peridomiciles. Currently, it is the species most frequently captured by the Chagas Disease Control Program in Brazil. For this reason, population genetic studies attract great interest, as they can provide further information about the dispersal and household invasion processes of this species. In the absence of suitable markers, the objective of this study was to test the cross amplification of microsatellite primers.
Findings
23 primers were tested for microsatellite loci already described for other species of the genus Triatoma sp. Forty four specimens of T. sordida captured in the north of Minas Gerais were used to validate the use of standardized loci for population genetic analyses. It was possible to amplify 10 of the 23 loci tested for T. sordida.
Conclusions
This is the first study that provides 10 microsatellite markers for population analysis of this triatomine species. Cross-amplification of primers can be used among other phylogenetically related species whose loci are already available for study.
Similar content being viewed by others
Findings
Background
Triatoma sordida (Stål, 1859) is the triatomine species most frequently captured by Brazil’s Chagas Disease Control Program [1] and in neighboring countries [2]. It occurs in Bolivia, Paraguay, Argentina and Uruguay [2], and Brazilian Cerrado [1]. This vector has broad ecological valence, and so it can live in several ecotopes and use different food sources [3]. It shows high rates of active dispersal [4], and it can also be passively introduced into artificial environments, possibly in firewood piles transported from forests to households, or from one household to another [5], or attached to bird feathers [3]. In an artificial environment, T. sordida is often associated with peridomiciles; in addition, it uses birds as its preferred hosts. Thus, this vector has secondary epidemiological importance [1].
Microsatellites are established as a valid technique to study diversity for more than a decade. Because they have highly polymorphic loci, they are widely used to investigate the genetic structure of natural populations. These genetic markers possess high reproducibility, multiallelic nature, codominant inheritance, abundance and wide distribution throughout the genome [6].
The use of microsatellites has not been widely applied in triatomines yet, but they are very sensitive markers and yield promising results in the population analyses of this subfamily. Microsatellite markers have been identified and characterized for the following triatomines: Rhodnius pallescens [7], Triatoma dimidiata [8], Triatoma infestans [9, 10], Rhodnius prolixus [11, 12], Triatoma pseudomaculata [13] and Triatoma brasiliensis [14]. Microsatellites described for particular species may sometimes be used to characterize others, e.g., markers for R. prolixus that are used for other species of the genus. However, this requires specific research that is certainly associated with the degree of similarity among species [11].
The analysis of gene flow between populations of the wild environment, intradomiciles and peridomiciles can help understand the factors that favor the infestation/reinfestation of the household, and hence provide guidelines for Chagas disease control programs [15, 16]. Considering this fact, the lack of microsatellite primers described for T. sordida, and also the great effort required for identifying and characterizing such markers, the objective of this study was to test the amplification of microsatellite loci in T. sordida using primers described for other triatomine species.
Methods
Primers were tested for 23 loci already described for other species of the genus Triatoma: T. dimidiata [8], T. infestans [10], T. pseudomaculata [13] and T. brasiliensis [14]. The insects used in the tests were provided from a mixed colony maintained in the insectarium of the Laboratory of Triatomines and Chagas Disease Epidemiology at the René Rachou Research Center. They were originally captured in several northern regions of Minas Gerais. Genomic DNA was extracted from one of the legs of three T. sordida adult specimens using the Wizard Genomic DNA Purification Kit (Promega). Quantitation of the DNA was performed in a NanoDrop® ND-1000 spectrophotometer, and the material was kept at −20 °C until processing.
Polymerase chain reactions (PCR) were performed in a final volume of 10 μL containing: 1 unit of Taq DNA Polymerase, Recombinant (Invitrogen), 1x buffer, 1.5 mM or 3 mM of MgCl2, 1 mM of dNTP, 5 pmoles of each primer, 2 ng of DNA and ultrapure water. Reactions were performed in a Eppendorf Mastercycles® Gradient thermocycler with the following cycle: initial denaturation at 94 °C for five minutes, 30 cycles at 94 °C for 30 s, temperature gradient dependent on annealing temperature in the primer description (±5 °C) for 30 s and 72 °C for 30 s, followed by final extension at 72 °C for three minutes. The amplified products were visualized in a polyacrylamide gel at 8 % in the mini-gel system (BIO-RAD), stained with 0.2 % silver nitrate.
To determine the size of the loci, new PCRs were performed with forward primers tagged with bioluminescent probe. The PCR products were diluted at 1:10 in ultrapure water and genotyped in a MEGABace (Amersham Biosciences) sequencer. The size of the PCR products was estimated in comparison with a standard size marker (ET-400, GE Health Care), and the genotypes were read using the software Fragment ProfilerTM.
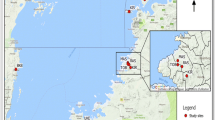
Specimens from the municipality of Coração de Jesus (16°41′15″S, 44°18′45″W), in the north of Minas Gerais, were used to test the use of standardized loci for T. sordida. These specimens were captured by agents of the Chagas Disease Control Program and not part of the mixed colony used for the selection of microsatellites. Forty four specimens of T. sordida were caught in the peridomicile of five neighboring localities: seven in Barriguda, nine in Boa Vista, 10 in Bom Jesus, 10 in Jataí I, and eight in Jataí II.
DNA was extracted from the wing muscle of adult insects [16]. Each muscle was homogenized individually in 100 μL of 1X STE solution (0.01 M NaCl, 0.1 M Tris–HCl and 1 M EDTA), incubated at 90°C for 10 min, and centrifuged at 13,000 rpm for one minute, and the supernatant was recovered. The quantitation and storage of DNA were carried out as described hereinabove.
The samples underwent standard PCR, the annealing temperature and the amount of MgCl2 for each primer are described in Table 1. The primers were tagged with bioluminescent probe; genotyping was also performed with standard conditions. For each locus, calculations were made of a number of alleles using the software Arlequin 3.1.
Results and discussion
Ten of the 23 pairs of tested primers showed satisfactory amplification for T. sordida in specific conditions. The size of the alleles obtained was similar to the original descriptions, except for loci Tb830 and Tp20, which were smaller for T. sordida (Table 1).
The number of alleles per locus ranged from 1 (TDMS4) to 12 (Tb 8124) with a mean of 6.5. Only TDMS4 locus showed no heterozygous individuals (Table 1).
Harry et al. [11] considered that cross-species amplification should be used with caution because microsatellite null alleles may occur. Their presence should be considered when population analyses are performed, as they may underestimate population diversity [17]. This study could validate the use of the 10 microsatellite markers.
Some of the primers described were tested for cross amplification in other species [10, 11, 13, 14], including T. sordida [9]. However, this is the first study that provides microsatellite markers for the population analysis of this triatomine species. Cross-amplification of primers among species was a low-cost, effective strategy which was faster than isolation, identification, and development of primers. Thus, it can be used among other phylogenetically related species whose loci are already available for study.
Due to the high sensitivity of the microsatellite markers, this study provides a new tool for the assessment of gene flow between populations of T. sordida also in microgeographic scale.
References
Gurgel-Gonçalves R, Galvão C, Costa J, Peterson AT. Geographic distribution of Chagas disease vectors in Brazil based on ecological niche modeling. J Trop Med. 2012;2012:1–15.
Moren AR, Carcavallo RU. Ecologic approach of chagas’ disease epidemiology. In: Carcavallo RU, editor. Atlas of chagas’ disease vectors in the americas, vol. III. 1st ed. Rio de Janeiro: Editora Fiocruz; 1999. p. 981–1016.
Forattini OP, Ferreira OA, Rocha E, Silva EO, Rabello EX. Aspectos ecológicos da tripanossomíase americana. VII-Permanência e mobilidade do Triatoma sordida em relação aos ecótopos artificiais. Rev Saude Publica. 1975;9:467–76.
Forattini OP, Ferreira OA, Silva EO d R e, Rabello EX. Ecological aspects of South American trypanosomiasis. VI. Persistence Triatoma sordida after environmental alterations and its possible relation to the dispersion of the species. Rev Saude Publica. 1974;8 Suppl 3:265–82.
Forattini OP, Rocha E, Silva EO, Ferreira OA, Rabello EX, Pattoli D. Aspectos ecológicos da Tripanossomíase Americana: III – Dispersão local de triatomíneos, com especial referência a Triatoma sordida. Rev Saude Publica. 1971;5:193–205.
Schlotterer C. The evolution of molecular markers- just a matter of fashion. Nat Rev Genet. 2004;5:63–9.
Harry M, Poyet G, Romana CA, Solignac M. Isolation and characterization of microsatellite markers in the Chagas disease vector Rhodnius pallescens. Mol Ecol. 1998;7:1784–6.
Anderson JM, Lai JE, Dotson EM, Cordon-Rosales C, Ponce C, Norris D, et al. Identification and characterization of microsatellites markers in the Chagas disease vector Triatoma dimidiata. Infect Genet Evol. 2002;1:243–8.
García BA, Zheng LO, de Rosas AR P, Segura EL. Isolation and characterization of polymorphic microsatellite locus in the Chagas disease vector Triatoma infestans (Hemipetera: Reduviidae). Mol Ecol Notes. 2004;4:568–71.
Marcet PL, Lehmann T, Groner G, Gurtler RE, Kitron U, Dotston EM. Identification and characterization of microsatellite markers in the Chagas disease Triatoma infestans (Heteroptera: Reduviidae). Infect Genet Evol. 2006;6:32–7.
Harry M, Roose CL, Vautrin D, Noireau F, Romana CA, Solignac M. Microsatellite markers from the Chagas disease vector, Rhodnius prolixus (Hemiptera, Reduviidae) and their applicability to Rhodnius species. Infect Genet Evol. 2008;8:381–5.
Fitzpatrick S, Watts PC, Feliciangeli MD, Miles MA, Kemp SJ. A panel of ten microsatellite loci for the Chagas disease vector Rhodnius prolixus (Hemiptera: Reduviidae). Infect Genet Evol. 2009;9(2):206–9.
Harry M, Dupont L, Romana C, Demanche C, Mercier A, Livet A, et al. Microsatellite markers in Triatoma pseudomaculata (Hemiptera, Reduviidae, Triatominae), Chagas’ disease vector in Brazil. Infect Genet Evol. 2008;8:672–5.
Harry M, Dupon TL, Quartie RM, Diotaiuti L, Walter A, Romana C. New perspective for population genetics of Chagas disease vectors in the Northeastern Brazil: isolation of polymorphic microsatellite markers in Triatoma brasiliensis. Infect Genet Evol. 2009;9:633–7.
Marcet PL, Mora MS, Cutrera AP, Jones L, Gürtler RE, Kitron U, et al. Genetic structure of Triatoma infestans populations in rural communities of Santiago del Estero, northern Argentina. Infect Genet Evol. 2008;6:835–46.
de Rosas AR P, Segura EL, García BA. Microssatellites analysis of genetic structure in natural Triatoma infestans (Hemiptera: Reduviidae) populations from Argentina: its implication in assessing the effectiveness of Chagas disease vector control programmes. Mol Ecol. 2007;16:1401–12.
de Queiroz BV, Coutinho-Abreu IV, Sonoda IV, Melo MA, de Andrade PP, JA d c, et al. Genetic structure of natural populations of the sandfly Lutzomyia longipalpis (Diptera: Psychodidae) from the Brazilian region. Acta Trop. 2006;98(1):15–24.
Acknowledgements
We are grateful to Dr. Marcela Lencine Ferraz and Dr. Bernardino Vaz de Melo for aiding in the collection of triatominae. We would also like to thank Centro Nacional de Ciência e Tecnologia (CNPq), Secretaria de Vigilância em Saúde/Brazil (SVS), Ministério da Saúde/Brazil, World Health Organization (WHO), and Fundação de Amparo à pesquisa de Minas Gerais (FAPEMIG) for their financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CJB participated in the design and standardized the study, data analysis, performed the statistical analysis, and drafted the manuscript. GCDP participated in the design, data analysis, and manuscript review. LSD, PFS, and ACLR participated in the samples preparation and processing. LSD also assisted in data analysis. LD conceived and coordinated the study, participated in the design of the study and manuscript review. All the authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Belisário, C.J., Pessoa, G.C.D., dos Santos, P.F. et al. Markers for the population genetics studies of Triatoma sordida (Hemiptera: Reduviidae). Parasites Vectors 8, 269 (2015). https://doi.org/10.1186/s13071-015-0879-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-0879-1