Abstract
Background
Layered double hydroxides (LDHs) have received widespread attention for their potential applications in catalysis, polymer nanocomposites, pharmaceuticals, and sensors. Here, the mechanism underlying the physiological effects of Mg–Fe layered double hydroxide nanoparticles on the marine bacterial species Arthrobacter oxidans KQ11 was investigated.
Results
Increased yields of marine dextranase (Aodex) were obtained by exposing A. oxidans KQ11 to Mg–Fe layered double hydroxide nanoparticles (Mg–Fe-LDH NPs). Furthermore, the potential effects of Mg–Fe-LDH NPs on bacterial growth and Aodex production were preliminarily investigated. A. oxidans KQ11 growth was not affected by exposure to the Mg–Fe-LDH NPs. In contrast, a U-shaped trend of Aodex production was observed after exposure to NPs at a concentration of 10 μg/L–100 mg/L, which was due to competition between Mg–Fe-LDH NP adsorption on Aodex and the promotion of Aodex expression by the NPs. The mechanism underling the effects of Mg–Fe-LDH NPs on A. oxidans KQ11 was investigated using a combination of physiological characterization, genomics, and transcriptomics. Exposure to 100 mg/L of Mg–Fe-LDH NPs led to NP adsorption onto Aodex, increased expression of Aodex, and generation of a new Shine-Dalgarno sequence (GGGAG) and sRNAs that both influenced the expression of Aodex. Moreover, the expressions of transcripts related to ferric iron metabolic functions were significantly influenced by treatment.
Conclusions
These results provide valuable information for further investigation of the A. oxidans KQ11 response to Mg–Fe-LDH NPs and will aid in achieving improved marine dextranase production, and even improve such activities in other marine microorganisms.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
Nanotechnology is a transformative tool that can be used to develop and enhance high-value products from renewable and biocompatible raw materials [1]. Nanoparticles (NPs) have attracted considerable interest due to their unique optical, electronic, and magnetic characteristics relative to their bulk counterparts [2]. Moreover, NPs are widely used in commercial products owing to their versatile properties, including surface areas, particle sizes, and quantum effects [3]. Layered double hydroxide (LDH) NPs, also known as anionic clay or hydrotalcite, are a family of inorganic lamellar materials with positively charged brucite-like layers comprising mixed metal hydroxides that are defined by the general formula [M II1− X M III X (OH)2]X+(An−)X/n·mH2O. Here, MII is a divalent cation, MIII is a trivalent metal cation, x is the molar ratio of the trivalent cation [MIII/(MIII + MII)], and An− is a gallery anion with charge n [4,5,6]. LDHs have received widespread attention in diverse applications, including in catalysis [7], polymer nanocomposites [8], pharmaceuticals [9, 10], and sensors [11]. Concomitantly, environmental pollution has emerged as an important problem over the last couple of decades, and interest is growing in using LDHs to remove environmental contaminants (e.g., heavy metals, pesticides, and polycyclic aromatic hydrocarbons) [12,13,14,15]. The large surface areas of LDHs play a vital role in enhancing the kinetics of electrochemical reactions and providing a large number of active sites for desired electrochemical reactivities [16].
In addition to the above, the investigation of LDH NP interactions with bacteria is of increasing interest. Numerous studies have shown that NPs can improve antimicrobial, anticorrosion, and antitumor functionalities through silver NPs [17], copper NPs [1, 18], and LDH NPs [19]. Moreover, other studies have shown that NPs can improve the growth of bacterial cells and their production of metabolites [20,21,22]. Specifically, NP aggregates can attach to and/or entrap cells, thereby impacting their cellular functions. It should also be noted that variation exists in the metabolite production by different microorganisms, including Escherichia coli, Bacillus, Bacillus subtilis, and Nocardiopsis sp., as indicated by their different production capacities and qualities [20, 23]. Al-Zn-LDH and Mg–Al-LDH [18, 24] LDHs have been intensively studied recently, while Mg–Fe-LDH has been much less investigated [6]. However, Mg–Fe-LDH NPs, which have been trademarked as Alpharen and Fermagate, have been intensively investigated in animal and clinical trials in the treatment of hyperphosphatemia in hemodialysis patients. Such studies have provided strong evidence of their high phosphate removal efficiency and biocompatibility [25, 26]. Moreover, increasing evidence indicates that Al can exert neurological, skeletal, and hematological toxicity. Consequently, the development of Al-free LDHs capable of maintaining highly efficient gene delivery has become increasingly desirable [26].
Few investigations have been conducted to evaluate the influence of LDHs on marine microorganisms. Indeed, investigation of LDH toxicity to microorganisms has been almost entirely conducted on terrestrial microorganisms, including Pseudomonas aeruginosa, Staphylococcus aureus, B. subtilis, and others [27,28,29]. However, most of the toxicities involved cell damage after analysis, which is likely resultant from low salinity tolerance of terrestrial microorganisms relative to marine microorganisms. It is well documented that marine microorganisms have high salt tolerance, hyperthermostability, barophilicity, alkali resistance, and low optimal growth temperatures. In addition, the intercalated molybdate anion can slowly diffuse out of the inner structure of LDHs in a controlled manner, resulting in relatively long-lived corrosion inhibition effects in marine anticorrosion applications [18].
Dextranases have drawn considerable attention due to their high potential for application in various fields, including in medical, dental, and sugar industries [30,31,32,33,34,35,36]. Dextranases (α-1,6-D-glucan 6-glucanohydrolase; EC 3.2.1.11) hydrolyze dextran to oligosaccharides at the α-1,6 glucosidic bond and are members of the glycoside hydrolase families (GH) 49 and 66 based on amino acid sequence homology [32, 37,38,39]. A. oxidans KQ11 was previously isolated by our research group from the Yellow Sea in the Lianyungang coastal region of China and produces dextranase (Aodex, Protein Data Bank code 6NZS) [36]. Here, we present a systematic study of the effects of repetitive dosing of various concentrations of Mg–Fe-LDH NPs on the marine bacterium A. oxidans KQ11 and its ability to produce Aodex. Transcriptional regulation is the mechanistic basis for bacterial growth and metabolism, and genome-wide transcriptional profiling can improve our understanding of the mechanisms underlying physiological changes [40, 41]. Consequently, transcriptional and genomic profiling was used to evaluate the mechanisms underlying variation in Aodex production following Mg–Fe-LDH NP exposure.
Results
Bacterial growth and Aodex production by A. oxidans in the presence of Mg–Fe-LDH NPs
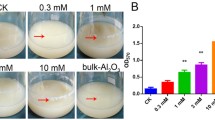
To determine the influence of Mg–Fe-LDH NPs on A. oxidans KQ11 growth and Aodex production, these properties were analyzed in the presence of varying concentrations of Mg–Fe-LDH NPs. When Mg–Fe-LDH NPs were added to bacterial cultures, statistically different levels of bacterial growth were not observed after 32 h (Fig. 1). This result was evident even at higher Mg–Fe-LDH NP concentrations (100 mg/L), implying that Mg–Fe-LDH NPs (at concentrations ≤ 100 mg/L) did not result in obvious growth effects on A. oxidans KQ11.
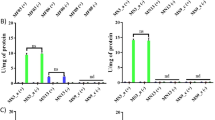
In contrast, enzyme production by A. oxidans KQ11 changed as a result of exposure to Mg–Fe-LDH NPs (Fig. 2A). Specifically, when exposed to 10 μg/L Mg–Fe-LDH NPs over 32 h, Aodex production was similar to that of the control and peak enzyme production occurred at 28 h, which was consistent with previous results [37]. The activities of Aodex upon exposure to 10 μg/L Mg–Fe-LDH NPs and the control were 3.69 U/mL and 4.03 U/mL, respectively. Exposure to increasing Mg–Fe-LDH NPs concentrations in the range of 10 μg/L to 1 mg/L resulted in decreased enzyme production, with Aodex production exhibiting the lowest activity after exposure to 1 mg/L Mg–Fe-LDH NPs. Peak enzyme production after exposure to Mg–Fe-LDH NPs at concentrations of 10 μg/L, 100 μg/L, and 1 mg/L was 3.69 U/mL at 28 h, 3.45 U/mL at 30 h, and 2.36 U/mL at 28 h, respectively. As increasing exposure to Mg–Fe-LDH NPs concentrations occurred beyond 1 mg/L, the overall trend of enzyme production began to rapidly increase. Indeed, enzyme production after exposure to 10 mg/L Mg–Fe-LDH NPs was similar to that after exposure to 100 μg/L Mg–Fe-LDH NPs, with a peak enzyme production of 3.43 U/mL at 30 h. Moreover, enzyme production was highest over the examined concentration range (10 μg/L to 100 mg/L) when exposed to an Mg–Fe-LDH NP concentration of 100 mg/L. Specifically, enzyme production in this treatment was 4.88 U/mL at 30 h, which was about 21.1% higher than the control, although peak enzyme production was delayed 2 h. Enzyme production after exposure to different Mg–Fe-LDH NP concentrations for 30 h was separately analyzed in order to directly compare enzyme production results (Fig. 2B), which also confirmed the above results. Briefly, enzyme production after exposure to Mg–Fe-LDH NPs concentrations in the range of 10 μg/L to 1 mg/L gradually declined, while enzyme production after exposure to Mg–Fe-LDH NPs concentrations in the range of 1 mg/L to 100 mg/L rapidly rebounded. The U-shaped trend of Aodex production resulted from the competition between Mg–Fe-LDH NP adsorption and the promotion of Aodex expression after exposure to NP treatment concentrations of 10 μg/L–100 mg/L.
Aodex generation after exposure of A. oxidans KQ11 to different concentrations of Mg–Fe-LDH NPs (A) and at 30 h (B). Columns with different letters indicate statistically significant differences (p < 0.05) between controls and treatments. The error bars represent standard deviations of averages (n = 3)
Characterization of bacterial cell morphology after exposure to Mg–Fe-LDH NPs
To gain further insight into the possible effects of Mg–Fe-LDH NPs on A. oxidans KQ11 cells, SEM, TEM, EDS, and EDS microscopy were used to examine A. oxidans KQ11 cells after exposure to 100 mg/L of Mg–Fe-LDH NPs. SEM (Fig. 3A, B) and TEM (Fig. 3C, D) images revealed that Mg–Fe-LDH NPs adhered to cellular membranes of A. oxidans KQ11 after washing cells three times, as noted by visible structures on the surface of A. oxidans KQ11 cells. However, alterations in A. oxidans KQ11 cell walls were not observed after exposure to 100 mg/L Mg–Fe-LDH NPs including a lack of surface disruptions, shrinkages, and irregularities.
Characterization of A. oxidans KQ11 cell structure after exposure to 100 mg/L of Mg–Fe-LDH NPs for 30 h using SEM (A, B), TEM (C, D), EDS (E), and EDS mapping (D–I). The yellow arrows show Mg–Fe-LDH NPs attached to cell surfaces (for interpretation of the color references to color in this figure legend, the reader is referred to the web version of this article)
To further determine the nature of the substances adsorbed on cell surfaces, the elemental composition of cell surfaces was investigated using EDS (Fig. 3E). These analyses indicated the presence of magnesium, but a lack of an iron signal, which may be due to the magnetic properties of iron and the low concentration used in these experiments that would render it difficult to detect using EDS. Consequently, EDS mapping was used to further detect the elements associated with cellular surfaces and further evaluate the presence of iron in association with cells (Fig. 3F). EDS mapping indicated the presence of phosphorus (Fig. 3G; green), magnesium (Fig. 3H; pink), and iron (Fig. 3I; yellow), thereby confirming that Mg and Fe were adhered to the surface of bacterial cells and implying that the structures on the surface of A. oxidans KQ11 were Mg–Fe-LDH NPs. The Aodex production experiments indicated that exposure to 100 mg/L of Mg–Fe-LDH NPs may influence cell membrane permeability, metabolite production, and/or gene expression in A. oxidans KQ11 cells, which would interact with cellular components to alter cellular processes. Mg–Fe-LDH NPs can likely penetrate cell membranes and reach cytosolic compartments due to their ability to dissolve slowly while releasing Mg2+ and Fe3+ ions. Nevertheless, understanding the exact mechanism by which Mg–Fe-LDH NPs improved Aodex production by A. oxidans KQ11 required further investigation.
Characterization of Mg–Fe-LDH NP endocytosis by A. oxidans KQ11
As described above, the primary influence of Mg–Fe-LDH NPs on enzyme production by A. oxidans KQ11 cells was through physical interactions with cells, and/or exposure to metal ions released from Mg–Fe-LDH. Thus, ICP-AES was used to measure the total intracellular metal ion content within A. oxidans KQ11 cells in order to identify the contribution of metal ions to the physiological influence of Mg–Fe-LDH NPs. The relative distribution and concentration of heavy metals within A. oxidans KQ11 cells of the Mg–Fe-LDH NP-treated bacterial cultures are shown in Fig. 4. Mg and Fe concentrations were all higher within A. oxidans KQ11 cells when exposed to Mg–Fe-LDH NP concentrations in the range of 10 μg/L to 100 mg/L. Furthermore, the concentration of Fe was significantly higher within A. oxidans KQ11 cells when exposed to 100 mg/L of Mg–Fe-LDH NPs than in cells of other treatment groups or the control group.
Transcriptional profiling of the A. oxidans KQ11 response to Mg–Fe-LDH NP exposure
The transcriptional response of A. oxidans KQ11 cells in response to Mg–Fe-LDH NPs was investigated with RNA-seq sequencing on the Illumina HiSeq platform. A total of 16.2 GBp of clean sequence read data were generated from the control and treatment samples. Specifically, gene-mapped transcript reads were generated three control (Control1, Control2, and Control3) and three Mg–Fe-LDH treatment (Mg–Fe-LDH1, Mg–Fe-LDH2, and Mg–Fe-LDH3) libraries, respectively (Additional file 1: Table S2). All unigenes were annotated using several databases including the CAZy, COG, GO, KEGG, NR, PFAM, and SwissProt databases (Additional file 1: Table S3). Non-redundant genes were obtained from all experimental samples and used as a transcriptome database to identify differentially expressed genes (DEGs, fold change > 2, p < 0.05) between controls samples and Mg–Fe-LDH-treated samples. These analyses indicated that 23 DEGs were up-regulated and 47 were down-regulated due to Mg–Fe-LDH exposure (Additional file 1: Table S4).
Discussion
Dextranases hydrolyze dextran to oligosaccharides at the α-1,6 glucosidic bond resulting in the production of isomaltose, isomaltotriose, small amounts of d-glucose, and traces of large oligomers as the primary products of the hydrolysis reaction. Previously, we isolated a dextranase from the marine bacterium A. oxidans KQ11 (Aodex; NCBI-n: JX481352.1) that was collected from Yellow Sea sediments near Lianyungang, China [37]. Aodex is active at low temperatures, is rapidly produced, and is stabile under alkaline conditions [37]. In addition, this enzyme derived from marine organisms has a characteristically high salinity tolerance and a low ideal temperature, conferring better application potential than homologous enzymes from terrestrial counterparts.
In this study, the growth of A. oxidans KQ11 after exposure to different concentrations of Mg–Fe-LDH NPs was not significantly influenced, while Aodex production was completely different among treatments. Specifically, enzyme production after exposure to Mg–Fe-LDH NPs at concentrations in the range of 10 μg/L to 1 mg/L gradually decreased with increasing concentration exposures. However, enzyme productions after exposure to Mg–Fe-LDH NPs concentrations in the range of 1 mg/L to 100 mg/L rapidly rebounded. To clarify the mechanism of Mg–Fe-LDH NP influence on A. oxidans KQ11 physiology, morphological and ultrastructural changes of cells were examined. These results indicated that Mg–Fe-LDH NPs could adhere to A. oxidans KQ11 cell membranes, but alterations in A. oxidans KQ11 cell walls were not observed, including a lack of surface disruptions, shrinkages, and irregularities after exposure to Mg–Fe-LDH NPs. Most marine microorganisms have unique physiological properties due to their unique environments compared to their terrestrial counterparts including high salt tolerance, hyperthermostability, barophilicity, alkali-resistance, and low optimum growth temperatures. Such characteristics could explain why A. oxidans KQ11 cell damage was not observed after exposures to such high concentration of Mg–Fe-LDH NPs. Concomitant to Aodex production rapidly increasing after exposure to 100 mg/L Mg–Fe-LDH, intracellular Fe concentrations were significantly higher in A. oxidans KQ11 after exposure to 100 mg/L Mg–Fe-LDH NPs when compared to the other treatment and control groups.
We previously described the structure of Mg–Fe-LDH NPs and their adsorption onto dextranase [6, 26]. Considering the gradual decrease in Aodex enzyme production after exposure to Mg–Fe-LDH NPs concentrations in the range of 10 μg/L to 1 mg/L, XRD (Additional file 1: Figure S1A) and FTIR (Additional file 1: Figure S1B) were used to investigate the adsorption onto Aodex by Mg–Fe-LDH NPs. The XRD spectra pattern of the Aodex/Mg–Fe-LDH biohybrid displayed characteristic diffraction peaks, with peak broadening and decreased intensity indicative of reduced crystallinity characteristics of the biohybrid. The FTIR spectra of the Mg–Fe-LDH NPs, Aodex, and Aodex/Mg–Fe-LDH NPs are shown in Additional file 1: Figure S1. The spectra exhibited broad and intense bands between 3900 and 2700/cm that were associated with the stretching of hydrogen-bonded hydroxyl groups from both the hydroxide layers and interlayered water molecules [24]. The shoulder located at about 3000/cm can be attributed to hydrogen bonding between water and anions located in the interlayer spacing including C–H, C–O–C, and C–O stretching bands [24, 42]. The bands at about 1650 and 1550/cm were assigned to the amide groups of amino acids, including C=O and N–H stretching bands. These results confirmed that Aodex was adsorbed on the surface of the Mg–Fe-LDH NPs, while also suggesting that the protein retained its secondary structure and did not denature. Thus, Aodex can adsorb on the surface of Mg–Fe-LDH NPs. Such characteristics provide a foundation for further utilization of Aodex, and especially in sugar industry applications, owing to the ease of release and effective removal of dextran generated during the production process. Previous studies have reported that LDHs can effectively immobilize numerous biomolecules or enzymes on their structures including laccase [43], polyphenol oxidase [44], urease [45], acid phosphatase–polyphenol oxidase [46], cytochrome c nitrite reductase [47], and horseradish peroxidase [48]. These characteristics enable their application in environmental pollutant monitoring including of cyanide, phenol derivatives, urea, azides, As (V), fluoride, nitrite, and H2O2.
While Mg–Fe-LDH NPs adsorbed the Aodex, Mg–Fe-LDH NPs also released Fe ions that could regulate Aodex production of A. oxidans KQ11. Thus, Mg–Fe-LDH NPs could contribute to more straightforward Aodex production via the above mechanism. Furthermore, this interaction could explain why Aodex production rapidly rebounded after exposure to 100 mg/L. To investigate the molecular mechanism underlying the regulation of Aodex productions by Mg–Fe-LDH, transcriptional profiling was conducted. A total of 4563 genes were expressed, and the expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced (FPKM) was used to identify genes that were differentially expressed. Using the criteria of a twofold change in expression and an FDR p value < 0.005, a total of 70 differentially expressed genes were identified. Of these, 23 genes were up-regulated and 47 were down-regulated in A. oxidans KQ11 after exposure to Mg–Fe-LDH NPs. The most important genes among those that were down-regulated included those that encoded siderophore synthetase components, iron complex transport system permease proteins, iron complex transport system ATP-binding proteins, NADPH-dependent ferric siderophore reductases, Fe-S cluster assembly ATP-binding proteins, and the Fe-S cluster assembly protein SufD, which are all directly related to inorganic ion transport and metabolism. The most important genes among those that were up-regulated included those that encoded N-acetylglucosamine-6-phosphate deacetylases, phosphotransferase system IIC components, ATPase components, predicted arabinose efflux permeases, sarcosine oxidase gamma subunits, 2,4-dienoyl-CoA reductases, formyltetrahydrofolate hydrolases, phosphotransferase system IIB components, 6-phosphogluconolactonase/glucosamine-6-phosphate isomerase/deaminases, sugar lactone lactonases, and malate synthases, which are all directly related to materials (carbohydrates, amino acids, and nucleotide) transport, metabolism, energy production and conversion, and signal transduction. The overall pattern of differential expression of transcription factor genes is shown in Fig. 5.
Changes in the gene expression of A. oxidans KQ11 cells induced by Mg–Fe-LDH treatment. A Significantly up-regulated genes are shown in red, down-regulated genes in blue, and those not exhibiting significant differences in expression are shown as black dots. The abscissa represents the fold changes in gene expression among different samples, and the ordinate represents the statistical significance of differences in expression change. B Heat map and clustering analysis of transcriptional profiles of genes encoding transcription factors. High expression levels are depicted in red, and low expression levels in blue. Clustering was conducted using log10 (FPKM + 1) values after normalization of expression values
GO analysis was also performed on the DEGs (Fig. 6). The DEGs all belonged to several categories, including cellular components, molecular functions, and biological processes. The majority of DEGs classified into the molecular function category were represented by those involved in catalysis, binding, and transportation activities. Among the DEGs classified into the cellular components category, membranes, cell parts, and membrane parts were prominently represented. Of the DEGs classified within the biological processes category, the vast majority were related to metabolic processes, cellular processes, localization, single-organism processes, cellular component organization or biogenesis, and biological regulation. The number of DEGs categorized as being involved in cellular components was lower than that of DEGs involved in molecular functions and biological processes (Fig. 6), which was consistent with the lack of observed cell damage in the experiments. Notably, 195 DEGs were classified as being involved in catalytic activity, while 70 were annotated as being involved in response to exposure, and 13 were annotated as being involved in ferric iron metabolism functions (particularly Fe3+ transport systems). These results provide a valuable framework for future studies of the response of A. oxidans KQ11 to Mg–Fe-LDH NP exposure.
The metabolic pathways coinciding with the DEGs were identified and analyzed using the KEGG database. The 16 most enriched pathways are shown in Fig. 7 and Table 1. Among the identified metabolic pathways, DEGs were primarily involved in the metabolism of biomolecules including carbohydrates and amino acids. Six DEGs were associated with ABC transporter pathways. Interestingly, DEGs involved in several carbohydrate transport and metabolism pathways including those associated with amino sugar and nucleotide sugar metabolism (3), phosphotransferase system (1), and glyoxylate and dicarboxylate metabolism (1) pathways were up-regulated in A. oxidans KQ11 after Mg–Fe-LDH NP treatment.
The Shine-Dalgarno sequence is a ribosomal binding site in bacterial and archaeal mRNA that is generally located around eight bases upstream of the start codon, AUG [49]. The sequence is also present in some chloroplast and mitochondrial transcripts. The RNA sequence helps recruit ribosome to mRNA in order to initiate protein synthesis and align the ribosome with the start codon. Once recruited, tRNA molecules can add amino acids sequentially, as dictated by the codons and moving downstream from the translational start site. The six-base consensus sequence is AGGAGG and AGGAGGU in E. coli, for example, although the subsequence GAGG dominates in E. coli virus T4 early genes [49]. The Shine-Dalgarno sequences of A. oxidans KQ11 RNA were predicted with RBSfinder after treatment with Mg–Fe-LDH NPs (Additional file 1: Table S5). A key difference was observed in the Shine-Dalgarno sequences with and without treatment, wherein the Shine-Dalgarno sequence of the Aodex (GeneID: KQ11_GM001677) position was 1,772,726 with the original start codon of CTA, but at a new starting position of 1,772,624 after treatment. Thus, the original sequence start coordinate moved upstream within the same reading frame. The new stop position also moved to 1,774,612 after treatment of A. oxidans KQ11 with Mg–Fe-LDH NPs and the pattern of the Shine-Dalgarno sequence changed to GGGAG, with the start codon changing from CTA to ATG. The transcriptional profile of A. oxidans KQ11 in the absence (control) and presence of Mg–Fe-LDH indicated that Aodex was up-regulated with an FDR p value of ~ 0.057. Thus, the GGGAG pattern of the Shine-Dalgarno sequence likely plays an important role in Aodex transcription and expression. Several studies have shown that base pairing between the Shine-Dalgarno sequence in mRNA and the 3′ end of 16S rRNA is critical for initiation of translation by bacterial ribosomes [50, 51]. Thus, mutations in Shine-Dalgarno sequence can reduce or increase translational responses in prokaryotes [52]. These changes are due to reduced or increased mRNA-ribosome pairing efficiencies, as evidenced by the restoration of translation by compensatory mutations in the 3′-terminal of 16S rRNA sequences.
Bacterial small RNAs (sRNA) are 50- to 500-nucleotide-long non-coding RNA molecules produced by bacteria that are highly structured and contain several stem loops [53]. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNAs or proteins and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress responses, and cell structures [54, 55]. Consequently, bacterial sRNAs exhibit a wide range of regulatory mechanisms. Generally, sRNAs bind to protein targets and modify the functions of bound proteins [56]. Alternatively, sRNAs can interact with mRNA targets and regulate gene expression by binding to complementary mRNAs and blocking translation, or by unmasking or blocking ribosome-binding sites. Many sRNAs are involved in the regulation of stress response [57] and are expressed under stress conditions such as cold shock, iron depletion, the onset of the SOS response, and sugar stress [58]. For example, the small RNA nitrogen stress-induced RNA 1 (NsiR1) is produced by cyanobacteria under nitrogen deprivation conditions [59]. In addition, cyanobacterial NisR8 and NsiR9 sRNAs could be involved in the differentiation of nitrogen-fixing cells (heterocysts) [60]. Novel non-coding sRNA transcripts in A. oxidans KQ11 intergenic regions that were expressed after Mg–Fe-LDH NP treatment were annotated by NR. A total of four sRNAs were identified, and their secondary structures were further predicted (Additional file 1: Table S6 and Fig. 8). These sRNAs may affect the expression of A. oxidans KQ11 intracellular proteins, and particularly the production of Aodex. Moreover, we hypothesize that these sRNAs are involved in the function of the riboswitches or the efficiency of Aodex production, which was up-regulated after Mg–Fe-LDH NP treatment (Fig. 9).
Conclusions
Our results indicated that the mechanism underlying the effects of Mg–Fe-LDH NPs on the physiology of the marine bacterium A. oxidans KQ11 could be related to the interaction of Fe3+, Shine-Dalgarno GGGAG sequences, and sRNAs (Fig. 8). As shown in Fig. 9, the proposed mechanism first involves adsorption of Mg–Fe-LDH NPs onto the surface of A. oxidans KQ11 cells, followed by release of Fe3+, which would impact carbohydrate transport and associated carbohydrate metabolisms. Secondly, the Shine-Dalgarno sequence is modified to GGGAG, which could play an important role in Aodex transcription and expression. Alternatively, the generation of sRNAs could interact with Aodex and further promote the expression of Aodex. This process would lead to constant Aodex expression with increasing concentrations of Mg–Fe-LDH NPs, while the adsorption of Mg–Fe-LDH NPs to enzymes would concomitantly gradually reach saturation. Such a mechanism would explain the U-shaped trend of Aodex production with increasing concentrations of Mg–Fe-LDH NPs. Nevertheless, abiotic stresses are undoubtedly complex in nature. However, understanding the full potential of biotechnological approaches can provide an important framework for improving enzyme production. Rapidly developing technologies including transcriptome profiling and nanotechnology provide promising future prospects for the development of designed enzymes that exhibit higher efficiency of natural resource utilization and improved productivity under stressful conditions.
The oceans cover more than three quarters of the Earth’s surface and are open ecosystems. The protection of marine environments and the reasonable exploitation and utilization of marine resources are vitally important to the sustainable development of human activities. Recent interest has grown for using LDHs to remove environmental contaminants. In this study, Mg–Fe-LDH NPs enhanced the production of Aodex by a marine bacterium. Furthermore, the mechanism underlying the influence of Mg–Fe-LDH NPs on the marine bacterium A. oxidans KQ11 (Fig. 1) was investigated using a combined approach of physiological characterization, genomics, and transcriptomics. These analyses indicated that cellular damage to marine A. oxidans KQ11 cells was not observed after Mg–Fe-LDH NP treatment. These results have an important practical significance wherein Mg–Fe-LDH NPs can be applied in the sustainable separation and extraction of marine resources without affecting marine microorganisms, even in marine ecosystems.
Materials and methods
A. oxidans KQ11 exposure to Mg–Fe-LDH NPs
Freshly grown bacterial colonies on solid nutrient agar medium were inoculated into 50 mL of A. oxidans KQ11 culture medium containing 1.0 g/L yeast extract, 5.0 g/L peptone, and NaCl at 4.0 g/L (pH 7.5). Growth was monitored with a UV–visible spectrophotometer at 600 nm until the optical density (OD) reached 0.8. Aliquots (10 µL) of the culture media were then further inoculated in 50 mL of freshly prepared nutrient broth medium containing 1.0 g/L yeast extract, 5.0 g/L peptone, NaCl 4.0 g/L, and 10 g/L dextran 20,000 (dextranase production medium, pH 7.5). The co-precipitation method was used for preparing Mg–Fe-LDH NPs (Additional file 1). In order to ensure Mg–Fe-LDH NPs with given concentrations were completely sterile for the growth and the enzyme production experiments, the Mg–Fe-LDH NP solutions were first sterilized by autoclaving. The Mg–Fe-LDH NPs were then sonicated at 20 kHz in a 100-W bath for 30 min at 25 °C. The Mg–Fe-LDH NP solutions were then sterilized with UV irradiation for 30 min. Subsequently, the Mg–Fe-LDH NPs were sonicated again at 20 kHz in a 100-W bath for 30 min at 25 °C before addition into A. oxidans KQ11 culture medium. Aliquots (50 mL) of A. oxidans KQ11 culture were exposed to various concentrations of Mg–Fe-LDH NPs (10 μg/L, 100 μg/L, 1 mg/L, 10 mg/L, and 100 mg/L) at the time of inoculation, and growth and enzyme production were monitored at 2-h intervals. All cultures were incubated at 30 °C in an orbital shaker incubator with shaking at 180 rpm and monitoring of bacterial growth in 2-h intervals via OD measurements at 600 nm using a microplate reader (Thermo Scientific™ Multiskan™ FC, Thermo Fisher Scientific Inc., Waltham, MA, USA). Controls consisting of medium without Mg–Fe-LDH NPs were conducted in parallel.
Dextranase activity assays
Aodex activity was measured using the DNS (3,5-dinitrosalicylic acid) method that is based on the reaction between sugars and the 3,5-dinitrosalicylic acid reagent, as described previously [37, 39]. Briefly, a mixture of 0.05 mL dextranase and 0.15 mL sodium acetate buffer (50 mM) containing 3% dextran 20,000 (pH 5.5) was incubated at 50 °C for 15 min. DNS [61] was added to the experimental and control mixtures to terminate the reactions, and 0.05 mL of enzyme was added to the control group. The mixture was boiled for 5 min, and then, 3 mL of distilled water was added. The absorbance of the mixture was then measured at 540 nm. One unit of dextranase activity was defined as the amount of enzyme that catalyzed the release of 1 μmol of isomaltose (measured as maltose) from dextran 20,000 in 1 min under the specified assay conditions [37]. To establish enzyme production curves, the thallus of the fermentation broth was removed by centrifugation after a specified time of aerobic fermentation and the liquid supernatant was filtered using an ultrafiltrate membrane (crude enzyme). The dextranase activities were then measured at specified times. For adsorbent experiments, the fermentation liquid supernatant recovered after centrifugation was used as the cell-free extract solution. The supernatant containing dextranase was then collected and purified. Briefly, dextranase was purified using a combination of ammonium sulfate fractionation and ion-exchange chromatography. SDS-PAGE and BD-SDS-PAGE (10% w/v SDS-PAGE with 0.5% Blue Dextran) analyses confirmed that the purified Aodex displayed a single band close to the expected molecular weight (66.2 kDa).
Ultrastructural observations and adsorbent characterization of A. oxidans KQ11
The Mg–Fe-LDH NP surface morphologies, physicochemical properties, elemental distribution, and interactions with bacterial cells were all investigated. The Mg–Fe-LDH NP solution was added to the A. oxidans KQ11 cultures after sonication (100 W, 20 kHz, 25 °C, 15 min) to obtain bacterial cultures containing Mg–Fe-LDH NPs at concentrations of 10 μg/L, 100 μg/L, 1 mg/L, 10 mg/L, and 100 mg/L. After 30 h of incubation, bacterial cells were collected by centrifugation of cultures for 10 min at 8000×g at 4 °C and then washed three times with sterile saline solutions. Scanning electron microscopy (SEM, Hitachi S-4000; Hitachi Instruments Inc., San Jose, CA, USA) and transmission electron microscopy (TEM, Hitachi HT7700; Hitachi Instruments Inc., San Jose, CA, USA) were then conducted on control cells and nanoparticle-treated cultures after suspension overnight in a phosphate-buffered saline (PBS) buffer with 2% glutaraldehyde. The pellet was then washed three times with the PBS buffer. A series of graded ethanol solutions (20%, 50%, 70%, 95%, and 100%) was used for dehydration over three exchanges consisting of 5 min each. STEM, energy-dispersive spectroscopy (EDS) and EDS mapping of A. oxidans KQ11 after treatment with 100 mg/L Mg–Fe-LDH NP were conducted using a JEOL 2100F microscope (JEOL, Tokyo, Japan).
Thirty milliliters of the bacterial culture suspensions containing 100 mg/L Mg–Fe-LDH NPs was collected after 30 h of exposure and centrifuged at 8000 rpm for 10 min. The residues were washed a third time with 50 mM PBS (pH 7.0), and cells were washed a third time with ultrapure water and then re-dissolved in 30 mL of ultrapure water. The supernatant following centrifugation was then used to determine the abundance of heavy metals adsorbed onto or into A. oxidans KQ11 cells. Heavy metal abundances were determined via inductively coupled plasma atomic emission spectrometry (ICP-AES, iCAP 6300, Thermo Fisher, USA). A. oxidans KQ11 suspensions without Mg–Fe-LDH NP exposure were used as controls.
Genomic and transcriptional analyses [62,63,64,65,66,67]
The whole genome of A. oxidans KQ11 was sequenced prior to transcriptome analyses. The A. oxidans KQ11 genome is not discussed in detail here, but annotation of the genome is provided in Additional file 1: Table S1.
Prokaryote mRNA sequencing on the Illumina HiSeq platform
RNA-Seq transcriptional profiling of A. oxidans KQ11 was conducted for cells in the absence (control) and presence of 100 mg/L Mg–Fe-LDH, with each group including three parallel replicates. Briefly, cells were harvested after exposure to Mg–Fe-LDH for 30 h and then centrifuged at 8000g (4 °C) for 10 min. Total cellular RNA of each sample was then extracted using a TRIzol reagent (Invitrogen)/RNeasy Mini Kit (Qiagen). Total RNA was quantified and quality-checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), NanoDrop spectrophotometer (Thermo Fisher Scientific Inc.), and 1% agarose gel electrophoresis. For subsequent library preparation, 1 μg of total RNA with an RIN value > 7 was used. Next-generation sequencing library preparations were constructed based on the manufacturer’s protocols (NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®).
Prior to sequencing, rRNA was depleted from total RNA using the Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina). The rRNA-depleted mRNA was then fragmented and reverse-transcribed. First-strand cDNA was synthesized using ProtoScript II Reverse Transcriptase with random primers and actinomycin D. The second-strand cDNA was then synthesized using a second-strand synthesis enzyme mix (including dACGTP/dUTP). The purified double-stranded cDNA was then cleaned using an AxyPrep Mag polymerase chain reaction (PCR) Clean-up kit (Axygen) followed by treatment with an End Prep enzyme mix to repair both ends of the fragments and add dA-tails in a single reaction, followed by T-A ligation to add adaptors to both fragment ends. Size selection of adapter-ligated DNA to recover fragments of ~ 360 bp length (approximate insert size of 300 bp) was then performed using an AxyPrep Mag PCR Clean-up kit (Axygen). The dUTP-containing second strand was then digested with a Uracil-Specific Excision Reagent (USER) enzyme (New England Biolabs). The DNA fragments of each sample were then amplified with PCR over 11 cycles using the P5 and P7 primers, with both primers carrying Illumina-specific sequences that can anneal to the sequencing flow cell and allow bridge PCR, in addition to a P7 primer carrying a six-base index to allow multiplexing. The PCR products were cleaned using an AxyPrep Mag PCR Clean-up kit (Axygen) and quality-checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) followed by quantification using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA).
Sequence libraries with different indices were multiplexed and sequenced on an Illumina HiSeq instrument according to manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was conducted with 2 × 150 paired-end (PE) chemistry, while image analysis and base calling were conducted using the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument.
Data analysis
Quality control
Filtering of poor-quality sequence reads including adapters, PCR primers, or fragments thereof, in addition to those with base quality scores < 20 were removed using Cutadapt (v1.9.1).
Mapping
Reference genome sequences and gene model annotation files were from genomes of close relatives of A. oxidans KQ11. Bowtie2 (v2.1.0) was then used to index the reference genome sequence. Clean sequence data were aligned and mapped to the reference genome using Bowtie2 (v2.1.0).
Expression analysis
Transcript sequence data in the FASTA format were first converted from gff annotation files and properly indexed. Then, using the file as a reference gene file, HTSeq (v0.6.1p1) was used to estimate gene expression levels from the paired-end clean sequence data.
Differential expression analysis
Differential expression analysis was conducted using the DESeq Bioconductor package that incorporates a model based on a negative binomial distribution of sequence abundances. After adjustment using Benjamini and Hochberg’s approach for minimizing the false discovery rate, a p value of < 0.05 was used to detect differentially expressed genes.
GO and KEGG enrichment analysis
The GO-TermFinder program was used to identify gene ontology (GO) terms among the annotated list of enriched genes exhibiting significantly different expression levels. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a collection of databases incorporating genomes, biological pathways, diseases, drugs, and chemical substances (http://en.wikipedia.org/wiki/KEGG). In-house scripts were used to identify significantly differentially expressed genes among different KEGG pathways. Lastly, the novel transcript prediction program Rockhopper uses a Bayesian approach to construct a transcriptome map including transcription start/stop sites for protein-coding genes and novel transcripts and was used to construct such a map using the transcriptional data generated here.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional file.
Abbreviations
- NPs:
-
nanoparticles
- LDH NPs:
-
layered double hydroxide nanoparticles
- Mg–Fe-LDH NPs:
-
Mg–Fe layered double hydroxide nanoparticles
- A. oxidans :
-
Arthrobacter oxidans
- Aodex:
-
A. oxidans KQ11 dextranase
- E. coli :
-
Escherichia coli
- B. subtilis :
-
Bacillus subtilis
- GH:
-
glycoside hydrolase families
- DNS:
-
3,5-dinitrosalicylic acid
- SEM:
-
scanning electron microscopy
- TEM:
-
transmission electron microscope
- PBS:
-
phosphate-buffered saline
- EDS:
-
energy-dispersive spectroscopy
- ICP-AES:
-
inductively coupled plasma atomic emission spectrometer
- GO:
-
gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- DEGs:
-
differentially expressed genes
- FPKM:
-
fragments per kilobase of transcript sequence per millions base pairs sequenced
- OD:
-
optical density
References
Rong-Mullins X, Winans MJ, Lee JB, Lonergan ZR, Pilolli VA, Weatherly LM, Carmenzind TW, Jiang L, Cumming JR, Oporto GS. Proteomic and genetic analysis of the response of S. cerevisiae to soluble copper leads to improvement of the antimicrobial function of cellulosic copper nanoparticles. Metallomics. 2017;9:1304–15.
Zhu X, Zhu L, Chen Y, Tian S. Acute toxicities of six manufactured nanomaterial suspensions to Daphnia magna. J Nanopart Res. 2008;11:67–75.
Baek S, Joo SH, Blackwelder P, Toborek M. Effects of coating materials on antibacterial properties of industrial and sunscreen-derived titanium-dioxide nanoparticles on Escherichia coli. Chemosphere. 2018;208:196–206.
Ding T, Lin K, Chen J, Hu Q, Yang B, Li J, Gan J. Causes and mechanisms on the toxicity of layered double hydroxide (LDH) to green algae Scenedesmus quadricauda. Sci Total Environ. 2018;635:1004.
Sideris PJ, Ulla Gro N, Zhehong G, Grey CP. Mg/Al ordering in layered double hydroxides revealed by multinuclear NMR spectroscopy. Science. 2008;321:113–7.
Ding Y, Liu L, Fang Y, Zhang X, Lyu M, Wang S. The adsorption of dextranase onto Mg/Fe-layered double hydroxide: insight into the immobilization. Nanomaterials. 2018;8:173.
Zhao Y, Jiao Q, Li C, Liang J. Catalytic synthesis of carbon nanostructures using layered double hydroxides as catalyst precursors. Carbon. 2007;45:2159–63.
Manzi-Nshuti C, Songtipya P, Manias E, Jimenez-Gasco MDM, Hossenlopp JM, Wilkie CA. Polymer nanocomposites using zinc aluminum and magnesium aluminum oleate layered double hydroxides: effects of the polymeric compatibilizer and of composition on the thermal and fire properties of PP/LDH nanocomposites. Polym Degrad Stab. 2009;94:2042–54.
Katharina L, Ping XZ, Qing Max LG. Layered double hydroxide nanoparticles in gene and drug delivery. Expert Opin Drug Deliv. 2009;6:907–22.
Soo-Jin C, Jin-Ho C. Layered double hydroxide nanoparticles as target-specific delivery carriers: uptake mechanism and toxicity. Nanomedicine. 2011;6:803–14.
Han J, Xu X, Rao X, Wei M, Evans DG, Duan X. Layer-by-layer assembly of layered double hydroxide/cobalt phthalocyanine ultrathin film and its application for sensors. J Mater Chem. 2011;21:2126–30.
Ambrogia V, Ciarnelli V, Nocchetti M, Rossi C. Effect of gliclazide immobilization into layered double hydroxide on drug release. Eur J Pharm Biopharm. 2009;73:285–91.
Koilraj P, Yu T, Sasaki K. Adsorption characteristics of arsenate on colloidal nanosheets of layered double hydroxide. Appl Clay Sci. 2016;134:110–9.
Li R, Wang JJ, Zhou B, Awasthi MK, Ali A, Zhang Z, Gaston LA, Lahori AH, Mahar A. Enhancing phosphate adsorption by Mg/Al layered double hydroxide functionalized biochar with different Mg/Al ratios. Sci Total Environ. 2016;559:121–9.
Peligro FR, Pavlovic I, Rojas R, Barriga C. Removal of heavy metals from simulated wastewater by in situ formation of layered double hydroxides. Chem Eng J. 2016;306:1035–40.
Baig N, Sajid M. Applications of layered double hydroxides based electrochemical sensors for determination of environmental pollutants: a review. Trends Environ Anal Chem. 2017;16:1–15.
Zhang H, Peng M, Cheng T, Zhao P, Qiu L, Zhou J, Lu G, Chen J. Silver nanoparticles-doped collagen–alginate antimicrobial biocomposite as potential wound dressing. J Mater Sci. 2018;53:14944–52.
Li W, Liu A, Tian H, Wang D. Controlled release of nitrate and molybdate intercalated in Zn-Al-layered double hydroxide nanocontainers towards marine anticorrosion applications. Colloid Interface Sci Commun. 2018;24:18–23.
Park DH, Cho J, Kwon OJ, Yun CO, Choy JH. Biodegradable Inorganic nanovector: passive versus active tumor targeting in sirna transportation. Angew Chem Int Ed. 2016;55:4612.
Mofradnia SR, Tavakoli Z, Yazdian F, Rashedi H, Rasekh B. Fe/starch nanoparticle—Pseudomonas aeruginosa: bio-physiochemical and MD studies. Int J Biol Macromol. 2018;117:51–61.
Fahim HA, Rouby WMAE, Elgendy AO, Khairalla AS, Naguib IA, Farghali AA. Enhancement of the productivity of the potent bacteriocin avicin A and improvement of its stability using nanotechnology approaches. Sci Rep. 2017;7:10604.
Alamdar N, Rasekh B, Yazdian F. Effects of Fe/SDS and Au nanoparticles on P. aeruginosa bacterial growth and biosurfactant production. IET Nanobiotechnol. 2018;12:520–5.
Current KM, Dissanayake NM, Obare SO. Effect of iron oxide nanoparticles and amoxicillin on bacterial growth in the presence of dissolved organic carbon. Biomedicines. 2017;5:55.
Nogueira KAB, Cecilia JA, Santos SO, Aguiar JE, Vilarrasa-García E, Rodríguez-Castellón E, Azevedo DCS, Silva IJ. Adsorption behavior of bovine serum albumin on Zn–Al and Mg–Al layered double hydroxides. J Sol-Gel Sci Technol. 2016;80:748–58.
McIntyre CW, Pai P, Warwick G, Wilkie M, Toft AJ, Hutchison AJ. Iron–magnesium hydroxycarbonate (fermagate): a novel non-calcium-containing phosphate binder for the treatment of hyperphosphatemia in chronic hemodialysis patients. Clin J Am Soc Nephrol. 2009;4:401–9.
Lu M, Shan Z, Andrea K, Macdonald B, Beale S, Curry DE, Wang L, Wang S, Oakes KD, Bennett C. Chemisorption mechanism of dna on mg/fe layered double hydroxide nanoparticles: insights into engineering effective SiRNA delivery systems. Langmuir. 2016;32:2659.
Bouaziz Z, Soussan L, Janot J-M, Jaber M, Amara ABH, Balme S. Dual role of layered double hydroxide nanocomposites on antibacterial activity and degradation of tetracycline and oxytetracyline. Chemosphere. 2018;206:175–83.
Djebbi MA, Elabed A, Bouaziz Z, Sadiki M, Elabed S, Namour P, Jaffrezic-Renault N, Amara ABH. Delivery system for berberine chloride based on the nanocarrier ZnAl-layered double hydroxide: physicochemical characterization, release behavior and evaluation of anti-bacterial potential. Int J Pharm. 2016;515:422–30.
Gayani B, Dilhari A, Wijesinghe GK, Kumarage S, Abayaweera G, Samarakoon SR, Perera IC, Kottegoda N, Weerasekera MM. Effect of natural curcuminoids‐intercalated layered double hydroxide nanohybrid against Staphylococcus aureus, Pseudomonas aeruginosa, and Enterococcus faecalis: a bactericidal, antibiofilm, and mechanistic study. MicrobiologyOpen. 2018;e00723.
Jiménez ER. Dextranase in sugar industry: a review. Sugar Tech. 2009;11(2):124–34.
Kaewprapan K, Inprakhon P, Marie E, Durand A. Enzymatically degradable nanoparticles of dextran esters as potential drug delivery systems. Carbohydr Polym. 2012;88:875–81.
Khalikova E, Susi P, Korpela T. Microbial dextran-hydrolyzing enzymes: fundamentals and applications. Microbiol Mol Biol Rev. 2005;69:306.
Keyes PH, Hicks MA, Goldman M, Mccabe RM, Fitzgerald RJ. 3. Dispersion of dextranous bacterial plaques on human teeth with dextranase. J Am Dent Assoc. 1971;82:136–41.
Purushe S, Prakash D, Nawani NN, Dhakephalkar P, Kapadnis B. Biocatalytic potential of an alkaliphilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manufacture. Bioresour Technol. 2012;115:2–7.
Wang X, Cheng H, Lu M, Fang Y, Jiao Y, Li W, Zhao G, Wang S. Dextranase from Arthrobacter oxydans KQ11-1 inhibits biofilm formation by polysaccharide hydrolysis. Biofouling. 2016;32:1223–33.
Ren W, Liu L, Gu L, Yan W, Feng Y, Dong D, Wang S, Lyu M, Wang C. Crystal structure of GH49 dextranase from Arthrobacter oxidans KQ11: identification of catalytic base and improvement of thermostability using semirational design based on B-factors. J Agric Food Chem. 2019;67:4355–66.
Wang D, Lu M, Wang S, Jiao Y, Li W, Zhu Q, Liu Z. Purification and characterization of a novel marine Arthrobacter oxydans KQ11 dextranase. Carbohydr Polym. 2014;106:71–6.
Ren W, Wang S, Lü M, Wang X, Fang Y, Jiao Y, Hu J. Optimization of four types of antimicrobial agents to increase the inhibitory ability of marine Arthrobacter oxydans KQ11 dextranase mouthwash. Chin J Oceanol Limnol. 2016;34:354–66.
Ren W, Cai R, Yan W, Lyu M, Fang Y, Wang S. Purification and characterization of a biofilm-degradable dextranase from a marine bacterium. Mar Drugs. 2018;16:51.
Victor C, Luca G, Karl K, Uwe S. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–40.
Zheng X, Wang J, Chen Y, Wei Y. Comprehensive analysis of transcriptional and proteomic profiling reveals silver nanoparticles-induced toxicity to bacterial denitrification. J Hazard Mater. 2017;344:291–8.
Gondim DR, Lima LP, de Souza MCM, Bresolin ITL, Adriano WS, Azevedo DCS, Silva IJ Jr. Dye ligand epoxide chitosan/alginate: a potential new stationary phase for human IgG purification. Adsorpt Sci Technol. 2012;30:701–11.
Mousty C, Vieille L, Cosnier S. Laccase immobilization in redox active layered double hydroxides: a reagentless amperometric biosensor. Biosens Bioelectron. 2007;22:1733–8.
Shan D, Cosnier S, Mousty C. Layered double hydroxides: an attractive material for electrochemical biosensor design. Anal Chem. 2003;75:3872–9.
Barhoumi H, Maaref A, Rammah M, Martelet C, Jaffrezic N, Mousty C, Vial S, Forano C. Urea biosensor based on Zn3Al–Urease layered double hydroxides nanohybrid coated on insulated silicon structures. Mater Sci Eng C. 2006;26:328–33.
Cosnier S, Mousty C, Cui X, Yang X, Dong S. Specific determination of As (V) by an acid phosphatase–polyphenol oxidase biosensor. Anal Chem. 2006;78:4985–9.
Chen H, Mousty C, Cosnier S, Silveira C, Moura J, Almeida M. Highly sensitive nitrite biosensor based on the electrical wiring of nitrite reductase by [ZnCr–AQS] LDH. Electrochem Commun. 2007;9:2240–5.
Shan D, Cosnier S, Mousty C. HRP wiring by redox active layered double hydroxides: application to the mediated H2O2 detection. Anal Lett. 2003;36:909–22.
Naglis M. Shine-Dalgarno sequence of bacteriophage T4: GAGG prevails in early genes. Mol Biol Rep. 2012;39:33–9.
Dahlberg AE. The functional role of ribosomal RNA in protein synthesis. Cell. 1989;57:525–9.
Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci. 1975;72:4734–8.
Johnson G, Widner W, Xin WN, Feiss M. Interference with phage lambda development by the small subunit of the phage 21 terminase, gp1. J Bacteriol. 1991;173:2733–8.
Vogel J, Wagner EGH. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol. 2007;10:262–70.
Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol. 2010;71:1–11.
Felden B, Vandenesch F, Bouloc P, Romby P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 2011;7:e1002006.
Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28.
Caswell CC, Oglesby-Sherrouse AG, Murphy ER. Sibling rivalry: related bacterial small RNAs and their redundant and non-redundant roles. Front Cell Infect Microbiol. 2014;4:151.
Nellen W, Hammann C. Small RNAs: analysis and regulatory functions. Berlin: Springer Science & Business Media; 2007.
Ionescu D, Voss B, Oren A, Hess WR, Muro-Pastor AM. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J Mol Biol. 2010;398:177–88.
Brenes-Álvarez M, Olmedo-Verd E, Vioque A, Muro-Pastor AM. Identification of conserved and potentially regulatory small RNAs in heterocystous cyanobacteria. Front Microbiol. 2016;7:48.
Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972;47:273–9.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Anders S, Huber W. Differential expression of RNA-Seq data at the gene level—the DESeq package. Heidelberg: European Molecular Biology Laboratory (EMBL); 2012.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2.
McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key R&D Program of China [Grant Number 2018YFC0311106], the Key Research and Development Program of Jiangsu [Social Development] [Grant Number BE2016702], and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
WR, ML, CW, and SW designed the experiments. WR and YD performed the experiments. WR, LG, WY, CW, and ML analyzed the data. SW and CW supervised the study and reviewed the manuscript. WR wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree to be published.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1.
The method of synthesis of Mg-Fe-LDH NPs, preparation of Aodex/Mg-Fe-LDH, and adsorbent characterization. Figure S1. Characterization of the adsorption of Aodex by Mg-Fe-LDH NPs. Table S1. Genome annotation of Arthrobacter oxidans KQ11. Table S2. for summary of raw and filtered reads; and Illumina transcriptome reads mapped to the A. oxidans KQ11 genes. Table S3. for transcriptome annotation of A. oxidans KQ11 response to Mg-Fe-LDH NPs. Table S4. for significantly differential expressed genes of A. oxidans KQ11 with the Mg-Fe-LDH NPS treatment. Table S5. for the Shine-Dalgarno sequences of A. oxidans KQ11 with the Mg-Fe-LDH NPs treatment. Table S6. for sRNA of the new transcripts in the A. oxidans KQ11 intergenic region with Mg-Fe-LDH NPs treatment were annotated by NR.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ren, W., Ding, Y., Gu, L. et al. Characterization and mechanism of the effects of Mg–Fe layered double hydroxide nanoparticles on a marine bacterium: new insights from genomic and transcriptional analyses. Biotechnol Biofuels 12, 196 (2019). https://doi.org/10.1186/s13068-019-1528-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-019-1528-2