Abstract
Background
The development of stratification tools based on the assessment of circulating mRNA of genes involved in the immune response is constrained by the heterogeneity of septic patients. The aim of this study is to develop a transcriptomic score based on a pragmatic combination of immune-related genes detected with a prototype multiplex PCR tool.
Methods
As training cohort, we used the gene expression dataset obtained from 176 critically ill patients enrolled in the REALISM study (NCT02638779) with various etiologies and still hospitalized in intensive care unit (ICU) at day 5–7. Based on the performances of each gene taken independently to identify patients developing ICU-acquired infections (ICU-AI) after day 5–7, we built an unweighted score assuming the independence of each gene. We then determined the performances of this score to identify a subgroup of patients at high risk to develop ICU-AI, and both longer ICU length of stay and mortality of this high-risk group were assessed. Finally, we validated the effectiveness of this score in a retrospective cohort of 257 septic patients.
Results
This transcriptomic score (TScore) enabled the identification of a high-risk group of patients (49%) with an increased rate of ICU-AI when compared to the low-risk group (49% vs. 4%, respectively), with longer ICU length of stay (13 days [95% CI 8–30] vs. 7 days [95% CI 6–9], p < 0.001) and higher ICU mortality (15% vs. 2%). High-risk patients exhibited biological features of immune suppression with low monocytic HLA-DR levels, higher immature neutrophils rates and higher IL10 concentrations. Using the TScore, we identified 160 high-risk patients (62%) in the validation cohort, with 30% of ICU-AI (vs. 18% in the low-risk group, p = 0.06), and significantly higher mortality and longer ICU length of stay.
Conclusions
The transcriptomic score provides a useful and reliable companion diagnostic tool to further develop immune modulating drugs in sepsis in the context of personalized medicine.
Similar content being viewed by others
Background
Sepsis is a complex life-threatening syndrome caused by a dysregulated host response to infection [1]. The primary inflammatory response is often followed by a complex and protracted immunosuppressive response affecting both the innate and adaptive components of immunity [2]. This post-septic immune suppression is associated with a higher risk of secondary infections, in the forefront of which are the intensive care unit (ICU)-acquired pneumonia [3]. However, a growing body of data suggest the existence of an important heterogeneity in the type and intensity of the immune response during post-septic immune suppression [4, 5]. Such heterogeneity is also retrieved at the patient level, sepsis from pulmonary origin being associated with a higher risk of subsequent ICU-acquired pneumonia when compared to sepsis from another origin [6]. Furthermore, the recent COVID-19 (Coronavirus disease 2019) pandemic has highlighted that the type of pathogen also influences the risk of ICU-acquired infection, the SARS-CoV-2 infected patients harboring a higher susceptibility to secondary pneumonia when compared to patients with flu or severe bacterial pneumonia [7, 8]. In this context, it is crucial to develop tools that allow for patient stratification according to their risk of ICU-acquired infections.
In this setting, transcriptomic approaches are promising for relevant informing stratification strategies in sepsis late phase. In a prospective cohort of septic patients, blood gene expression at the onset of ICU-acquired infections showed reduced expression of genes involved in gluconeogenesis and glycolysis [3]. In ICU patients with ventilator-associated pneumonia (VAP), authors reported transcriptomic depression of genes involved in the immunological synapse in the blood [9]. However, almost all studies investigate the transcriptomic response during the ICU-acquired infection rather than identify patients at risk to develop further infection. The recent development of multiplex molecular platforms such as the FilmArray System (bioMérieux) allows for rapid and reliable evaluation of the transcriptomic response of patients during nosocomial infections on the basis of the expressions of several genes involved in the pro- and anti-inflammatory response [10]. In this setting, we hypothesize that an analytical approach of gene expression, considering that each gene is mutually independent of the others and that they have a similar importance in the models, is relevant. We report here the diagnosis performance of a transcriptomic score performed between day 5 and day 7 after ICU admission that identify a subgroup of critically patients who are likely to exhibit poor clinical outcomes, including higher rates of ICU-acquired infection, longer ICU length of stay and higher mortality.
Methods
Patients and setting
In the training cohort, the patients and the data are abstracted from the previously published REALISM (REAnimation Low Immune Status Marker) cohort study [11]. Briefly, this monocenter observational cohort study included critically ill patients with sepsis, trauma and burn patients between December 2015 and June 2018. Inclusion criteria were: patients aged > 18 years, clinical diagnosis of sepsis as defined by the SEPSIS-3 consensus guidelines [1], severe trauma with injury severity score (ISS) > 15 or total burned surface area > 30%. Exclusion criteria were any of the following: presence of a preexistent condition or treatment that could influence patients’ immune status, pregnancy, institutionalized patients, inability to obtain informed consent. Whereas data and biological samples were collected at day 1 or 2 (D1-2), D3 or 4 (D3-4) and D5, D6 or D7 (D5-7), we focused on day 5–7. Longitudinal follow-up was performed for a period of 90 days. The IRB (Comité de Protection des Personnes Sud-Est II) approved the study (ref. 2015-42-2), and this study was registered at clinicaltrials.gov (NCT 02638779).
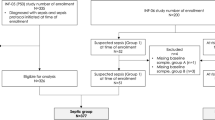
The validation cohort is derived from the MIPrea (Marqueurs Immunitaires Pronostiques en Réanimation) study [12] that enrolled patients aged > 18 years old with an expected length of stay > 2 days and SIRS criteria [13] between December 2009 and June 2011 in six French ICUs (approval CIC IRB #5044). This validation cohort consisted exclusively in patients with sepsis defined using the SEPSIS-2 definition [14]. In this study, we also analyzed biological data at day 5–7.
Intended management
Septic patients were treated according to the guidelines of the Surviving Sepsis Campaign guidelines [15]. Patients received intravenous broad-spectrum antibiotics, depending on the presumed site of infection, previous antibiotic treatment and known colonization with antibiotic-resistant bacteria. Antimicrobial treatment was deescalated to narrower spectrum after identification of the responsible pathogen. Source control measures, such as surgery or removal of infected devices, were applied when necessary.
After initial stabilization using the “Parkland formula” for fluid resuscitation [16], burn patients were evaluated quickly to assess the need for debridement, escharotomy and fasciotomy in case of compartment syndrome. Opioids were used for pain control and sedation in addition to hypnotics. Patients underwent several surgical procedures for cleaning, non-viable tissue removal, skin graft and or amputation according to local aspect.
Initial management of severe trauma aimed to control the post-traumatic hemorrhage, including surgical procedures for damage-control and medical treatment of coagulopathy, and to stabilize hemodynamics with vasopressors if required. Patients were intubated and mechanically ventilated if alteration of consciousness, for preventing hypoxia. A restricted strategy of blood transfusion was applied and targeted hemoglobin of 70 to 90 g/L Pain management was based on opioids administration, combined with regional anesthesia if adapted, or associated with hypnotics in sedated patients. Patients could undergo several surgical procedures according to location of trauma [17].
Definitions
Severity at admission was assessed by the Simplified Acute Physiology Score 2 and the Sequential Organ Failure Assessment (SOFA) scores in septic patients and using the ISS score in trauma patients [18,19,20]. ICU-acquired infections were defined as any new onset of probable or definite infection that developed after 48 h from ICU admission. Only the first episode of ICU-acquired infection was considered for analysis. ICU-acquired pneumonia was diagnosed according to the American Thoracic Society criteria [21]. Patients with clinically suspected ventilator-associated pneumonia were usually subjected to a tracheobronchial aspirate with semiquantitative cultures. Diagnosis of catheter-related bloodstream infection required the growth of the same pathogen from both peripheral blood and catheter tip cultures, or from blood cultures sampled from the catheter and from venous puncture with a differential time to positivity > 120 min. Urinary tract infections, mostly catheter related, were diagnosed upon the association of systemic manifestations of infection and positive urine bacterial culture at ≥ 105 CFU/mL. An independent adjudication committee composed of three clinicians not involved in study patients’ recruitment or care reviewed the infections.
Immunological measurements
The determination of the number of HLA-DR molecules per monocyte using the BD Quantibrite standardized method (HLA-DR: 340827; Quantibrite: 340495; Becton Dickenson, New Jersey, USA) was performed on fresh EDTA blood samples, within 3 h after collection. Plasma IL-10 level measurement was performed in ELISA using EDTA samples collected from patients.
Immune multiplex molecular tool
The multiplex molecular tool was performed as previously reported [10]. Briefly, one PAXgene blood RNA tube (PreAnalytix, Hilden, Germany) was sampled at each time point, stabilized for at least 2 h at room temperature after collection and frozen at − 80 °C following manufacturer’s recommendations. mRNA expression was quantified using the FilmArray instrument (bioMérieux) for automatic mRNA reverse transcription, amplification and further quantitative nested PCR of twenty-six genes (ADGRE3, ARL14EP, BPGM, C3AR1, CCNB1IP1, CD177, CD274, CD3D, CD74, CIITA, CTLA4, CX3CR1, GNLY, IFNG, IL10, IL1R2, IL1RN, IL7R, IP10/ CXCL10, MDC1, OAS2, S100A9, TAP2, TDRD9, TNF and ZAP70). Of note, those twenty-six genes were selected on the basis of the literature [12, 22,23,24,25,26,27] and preliminary analysis from the REALISM study [11].
Calculation of the transcriptomic score and statistical analysis
The normalized expression of individual transcripts of each gene evaluated by the multiplex molecular tool was first compared between healthy volunteers, patients with and without ICU-acquired infection. The receiver operator characteristics (ROC) curve for each gene to differentiate patient with and without ICU-acquired infection was then calculated. For genes with an area under the curve (AUC) > 0.70, we then calculated the optimal cut-point value as the value that maximizes the Youden’s index: sensitivity (%) + specificity (%) − 100. As sensitivity analysis, we additionally computed optimal cut-point using OOP (optimal operating point) method that is the value minimizing the distance from 100 of sensitivity and specificity: \(\sqrt {\left( {100 - sensitivity} \right)^{2} + \left( {100 - specificity} \right)^{2} }\) [28]. The cut-point value of each gene is then used to define a threshold value above or below which (depending on its increase or decrease in patients with ICU-acquired infection) each gene is allocated or not a point. The transcriptomic score was therefore the sum of all the values for a given patient.
Results
Training cohort
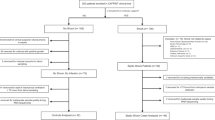
In the REALISM cohort, 324 patients had a blood sampling at day 5–7. Among them, 176 did not presented with an ICU-acquired pneumonia before day 5–7 blood sampling and were still in the ICU at that time point. Most patients included in this training set were admitted for trauma (n = 72, 41%) and sepsis (n = 68, 39%). The overall ICU mortality was 8%.
Overall, 47 patients presented at least one episode of ICU-acquired infection while 129 patients did not. Most ICU-acquired infections were ICU-acquired pneumonia (n = 21, 37%) (Fig. 1A). Patients who presented an ICU-acquired infection had more frequent diabetes mellitus (21% vs. 14%) and chronic pulmonary disease (19% vs. 9%) when compared to patients without ICU-acquired infections (Table 1). The median duration of mechanical ventilation was longer in patients with ICU-acquired infection when compared to patients without ICU-AI (15 days (95% IQR: 1–29) vs. 1 day (0–3), respectively).
Using our approach (Fig. 1C), among the 26 genes included in the multiplex molecular tool, 8 transcripts had an AUC value upper than 0.70 to classify patients with or without ICU-acquired infection, namely C3AR1, CD177, CX3CR1, IFNγ, IL1R2, S100A9, TDRD9 and ZAP70 (Fig. 2A). Normalized mRNA transcript expression was increased in patients with ICU-acquired infections except for CX3CR1, IFNγ and ZAP70 (Fig. 2B). The main diagnosis performances and cut-point value for each of the 8 transcripts are summarized in Table 2.
Gene’s identification process in the discovery cohort. A ROC (Receiver Operating Characteristics) curves with AUC (area under the curve) upper than 0.70 of several genes (C3AR1: Complement C3a Receptor 1, CD177: CD177 molecule, CX3CR1: C-X3-C motif chemokine receptor 1, IFNγ: Interferon gamma, IL1R2: Interleukin 1 receptor 2, S100A9: S100 calcium binding protein A9, TDRD9: Tudor domain containing 9, ZAP70: Zeta chain of T cell receptor-associated protein kinase 70), B Normalized RNA values for identified genes in patient with (red dots) or without (green) ICU-acquired infections (ICU-AI)
We built the transcriptomic score (TScore) using the threshold values obtained with the Youden technique on the ROCs for each transcript. For each gene analyzed individually, patients with a value above the threshold (for C3AR1, CD177, IL1R2, S100A9 and TDRD9) or below threshold (for CX3CR1, IFNγ and ZAP70) were assigned a point. Conversely, genes that do not meet these criteria were assigned a value of 0. The TScore, which varies between 0 and 8, is the sum of the values for each of the 8 genes (Fig. 3A). The AUC value of this TScore performed at day 5–7 is 0.86 (0.80–0.92) in distinguishing patients with or without at least one episode of ICU-acquired infection during their ICU stay (Fig. 3B). As sensitivity analysis, the OOP approach retrieved similar results but with lower ROC AUC for TScore (Additional file 1: Table S1 and Fig. S1).
Clinical outcomes of high- and low-risk patients in the discovery cohort using the TScore. A Heatmap of unsupervised hierarchical clustering in patients with or without ICU-acquired infections using individual gene score, B ROC (Receiver Operating Characteristics) curve with AUC (area under the curve) value of the ability of the TScore obtained at day 5–7 to distinguish between patients that will develop or not at least an ICU-acquired infection during their ICU stay, C Intensive care unit acquired infection (ICU-AI) proportion in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk), D Distribution of the type of ICU-AI in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk) (UTI: urinary tract infections), E Median and interquartile duration of intensive care unit length of stay in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk), F: intensive care unit mortality rate in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk)
Using the Youden index, the optimal value of the score is 2. We therefore defined a high-risk population of patients with a TScore upper to 2 and a low-risk population of patients with a TScore lower or equal to 2.
When applied to the whole population of the training set, patients with a TScore between 3 and 8 (n = 87, 49%) presented a higher proportion of ICU-acquired infection when compared to patients with a TScore lower or equal to 3 (49% vs. 4%, respectively) (Fig. 3C). In these high-risk patients, ICU-acquired infections were more frequently pneumonia (n = 19, 49%) when compared to low-risk patients (n = 2, 11%) (Fig. 3D). Furthermore, they experienced a longer median ICU length of stay (13 days, 25th–75th IQR: 8–30 vs. 7 days, 25th-75th IQR: 6–9, p < 0.001) when compared to patients in the low-risk TScore group (Fig. 3E). The ICU mortality in the high-risk TScore group was higher than the mortality of the patients in the low-risk TScore group (15% vs. 2%, p < 0.01) (Fig. 3F). All parameters are summarized in Table 3.
In the training set, a Wilcoxon–Mann–Whitney test was performed to determine potential relevant clinical and biological variables of interest for predicting the occurrence of ICU-acquired infections. Of the variables analyzed, only SOFA score at day 6 and T score showed a statistically different distribution between ICU-acquired infections and no ICU-acquired infections. In a multivariate model, the TScore remained independently associated with ICU-acquired infections occurrence (Table 4).
A type I ANOVA was applied, varying the order of introduction of the explanatory variables in the model, to determine the own contribution of the explanatory variables TScore and SOFA at day 6 to the minimization of deviance, as well as their joint contribution. The model thus created explains 25.4% of the total deviance, with 81% being explained by the TScore alone, 16.8% of the deviance explained by the TScore and the SOFA at day 6 together, and 1.7% by SOFA at day 6 alone. TScore therefore only slightly overlaps with information already explained by the main clinical variables in the REALISM database and the addition of such variables to TScore seems of little relevance in regard of the low proportion of deviance they explained in the model (Fig. 4A).
Statistical and immunological validity of the TScore in the discovery cohort. A picture representing the respective participation of each parameter in the deviance of the model and their overlap, B level of expression of several immunological parameters (monocytic HLA-DR, IL-10 and immature neutrophils proportion) measured in blood of patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk) and C clinical parameters and outcomes of patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk) sub-classified according to their subgroup
Finally, we investigated the expression level of several biomarkers previously reported to be associated with poor outcomes in ICU patients [29]. Interestingly, we found that the monocytic HLA-DR level of expression was decreased in high-risk patients. Similarly, the concentration of the anti-inflammatory cytokine IL10 and the immature neutrophils rate was increased in high-risk patients (Fig. 4B and Additional file 1: Table S2). Finally, when we apply our model to the training cohort according to the initial aggression (sepsis, burns, trauma and surgery), we find the same capabilities of the TScore to identify a group at high risk of complications (Table 5).
Validation cohort
For the validation cohort, we assessed the diagnosis performance of the TScore in septic patients from the MIPrea cohort. The proportion of high-risk patients with a TScore between 3 and 8 was higher in the MIPrea cohort when compared to the REALISM cohort (62% vs. 49%, respectively). High-risk patients were most severe at ICU admission, with a higher SOFA score (10, 25th–75th IQR: 7–13 vs. 8, 25th–75th IQR: 5–11) and higher rate of mechanical ventilation (87% vs. 75%) (Table 6).
ICU-acquired infections were more frequent in high-risk patients (30% vs. 18%, p = 0.06) (Fig. 5B), with a lower proportion of urinary tract infections (Fig. 5C). The median ICU length of stay was longer in high-risk patients (12 days, 25th–75th IQR: 8–20 vs. 9 days, 25th–75th IQR: 7–14, p = 0.003) (Fig. 5D). ICU mortality was also higher in high-risk patients (20% vs. 13%, p = 0.006) (Fig. 5E).
Clinical outcomes of high- and low-risk patients in the validation cohort using the TScore. A Intensive care unit acquired infection (ICU-AI) proportion in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk), B Distribution of the type of ICU-AI in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk) (UTI: urinary tract infections), C Median and interquartile duration of intensive care unit length of stay in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk), D intensive care unit mortality rate in patients with a TScore between 0 and 2 (low risk) or upper or equal to 3 (high risk)
Discussion
Two groups of patients with different risks of poor outcome, including ICU-acquired infections, mortality and length of ICU stay, were identified using a transcriptomic score based on the expression level of 8 genes. Relative to low-risk patients, patients in the high-risk group had an increased risk of ICU-acquired infections, even after adjustment for SOFA score, longer duration of ICU stay and higher mortality. High-risk assignment at day 5–7 was associated with lower mHLA-DR values and higher circulating IL10 values.
Owing to early identification of sepsis, rapid initiation of antibiotics and organ support therapies, many septic patients now survive the early phase of sepsis. However, they are therefore exposed to a higher risk of secondary infections that are associated with several markers of immune dysfunction such as downregulated monocytic HLA-DR expression [30]. Despite a robust pathophysiological rationale, several randomized clinical trials have failed in showing a beneficial effect of immune modulating therapies over the last decades [31]. One of the hypothesis for this failure relies on the important heterogeneity in septic patients and suggests to stratify patients on the basis of immune status in order to identify those who are more likely to benefit from these therapies [32]. Such stratification requires reliable, relevant, rapid and decision-making monitoring tools.
With this aim, assessment of the global differential gene expression within blood has provided encouraging results in the early phase of sepsis [27, 33, 34]. However, whether most of the studies fostered on mortality [23, 27, 33, 35], only a few genes were reported as relevant biomarkers to predict ICU-acquired infections [25]. Indeed, it is difficult to identify genes associated with a higher risk of ICU-acquired infection and several hypotheses can be formulated to explain it. First, blood cellular composition and gene expression vary rapidly over time depending on patient-related and management-related parameters. Second, the onset of sepsis remains unknown in most cases. Therefore, the simultaneous assessment of the expression of several genes appears as an interesting approach in order to pick up a signal whatever the time when the sample is performed [10]. To do so, we took advantages of the advent of multiplex molecular platforms such as the bioMérieux FilmArray System that allows the realization of rapid and reliable evaluation of the expressions of several genes. We performed our analysis between 5 and 7 after ICU admission, which accurately captures the impact of both the underlying condition and critical care interventions on the patient's immune status. This time frame also allows to shift away from the initially dysregulated immunity and reach the moment where some patients exhibit clear sign of return to homeostasis while others keep being dysregulated [36]. Furthermore, transcriptomic results were unaffected by the occurrence of ICU-acquired infections that were absent at this timepoint.
Over the last years, machine learning (ML) algorithms have gained increasing interest for classification, unsupervised clustering or dimensionality reduction tasks of large data sets. ML tools use data-driven algorithms and statistical models to analyze data sets and then, draw inferences from identified patterns or make predictions based on them. However, ML algorithms based on gene-expression performances in predicting ICU-acquired infections remain similar to usual severity scores [37]. These poor results could be related to the mismatch between two main assumptions of the ML algorithms and the characteristics of the clinical and biological data from which the algorithms are derived. First, ML algorithms assume the absence of covariate shift between training and test set. This assumption is frequently violated in ICU given the important heterogeneity in patient’s characteristics. Second, ML models intrinsically generate different features importance among dependent variables in order to fit as well as possible to the data. This assumption may be responsible for data overfitting and therefore, limit generalization of the models.
Based on these observations, we built the TScore, considering that 1/ each gene is independently associated with ICU-acquired infection and has an equal importance in the score without any weighting and 2/ that genes included in the score are mutually independent. This possibly allows for the capture of different pathophysiological mechanisms that would reflect an alteration of the immune system. This simplistic approach does not take into account the possible interactions between genes in signaling pathways that may be involved. However, given the differences among studies regarding the setting and the heterogeneity in septic patients, we believe this approach is appropriate in capturing relevant information in heterogeneous patients.
We found that the TScore was associated with the occurrence of ICU-acquired infections independently of the clinical variables usually associated with it. Furthermore, we found in high-risk patients biological features of immune suppression, with low levels of monocytic HLA-DR, high rate of immature neutrophils and high levels of the anti-inflammatory cytokine IL-10. These data suggest that the TScore is able to identify patients at risk of ICU-acquired infections based on circulating immune alterations. Therefore, our results suggest that this TScore could be used as a stratifying tool to identify patients likely to benefit from immune therapies. This approach could also allow to propose specific strategies for the prevention of ICU-AI, for instance by specifically monitoring microbiological carriage or by implementing dedicated prophylactic measures.
This score was build using a cohort of critically ill patients admitted in the ICU with various etiologies, including severe infections, burns, trauma and surgery. In line with previously published results, we postulated that immune alterations in circulating cells are shared among various medical conditions [11]. We then were able to validate this approach on a cohort of patients exhibiting exclusively a severe infection, which shows the robustness of the score.
This study acknowledges several limitations. First, we provided validation in an historical independent cohort and not in a prospectively collected cohort. Second, even if the results were repeated on a validation cohort, our results need to be evaluated in a large cohort and performed in a prospective way to confirm the performances of this test. Third, our results are based on whole-blood leukocyte populations that may differ among patients. Therefore, we cannot exclude that our results rather reflect differences in leukocytes populations rather than intracellular gene-expression differences. Fourth, the PCR multiplex was performed at day 5–7 after ICU admission. This limits the diagnostic performance of this test to the time period to which it is performed. The TScore must be validated and/or adapted at several time points, which should be possible thanks to the agility and plasticity of the platform.
Conclusions
Using a multiplex molecular platform, we built a transcriptomic score based on simultaneous assessment of the expression of eight genes at day 5–7 after ICU admission that identifies a subgroup of patients at high risk to develop ICU-acquired infections. The host response that is detected in high-risk patients is associated with known biomarkers of immune dysfunction and provide a useful and reliable companion diagnostic tool to further develop immune modulating drugs evaluation in sepsis.
Availability of data and materials
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Abbreviations
- AUC:
-
Area under the curve
- COVID-19:
-
Coronavirus disease 2019
- ICU-AI:
-
Intensive care unit acquired infections
- ML:
-
Machine learning
- PCR:
-
Polymerase chain reaction
- ROC:
-
Receiver operator characteristics
- SOFA:
-
Sequential organ failure assessment
- VAP:
-
Ventilator-associated pneumonia
References
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):801.
Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 2013;13(3):260–8.
van Vught LA, Klein Klouwenberg PMC, Spitoni C, Scicluna BP, Wiewel MA, Horn J, et al. Incidence, risk factors, and attributable mortality of secondary infections in the intensive care unit after admission for sepsis. JAMA. 2016;315(14):1469–79.
Llitjos JF, Bounab Y, Rousseau C, Dixneuf S, Rimbault B, Chiche JD, et al. Assessing the functional heterogeneity of monocytes in human septic shock: a proof-of-concept microfluidic assay of TNFα secretion. Front Immunol. 2021;12: 686111.
Avraham R, Haseley N, Brown D, Penaranda C, Jijon HB, Trombetta JJ, et al. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell. 2015;162(6):1309–21.
Llitjos JF, Gassama A, Charpentier J, Lambert J, de Roquetaillade C, Cariou A, et al. Pulmonary infections prime the development of subsequent ICU-acquired pneumonia in septic shock. Ann Intensive Care. 2019;9(1):39.
Llitjos JF, Bredin S, Lascarrou JB, Soumagne T, Cojocaru M, Leclerc M, et al. Increased susceptibility to intensive care unit-acquired pneumonia in severe COVID-19 patients: a multicentre retrospective cohort study. Ann Intensive Care. 2021;11(1):20.
Rouzé A, Martin-Loeches I, Povoa P, Makris D, Artigas A, Bouchereau M, et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: a European multicenter cohort study. Intensive Care Med. 2021;47(2):188–98.
Almansa R, Nogales L, Martín-Fernández M, Batlle M, Villareal E, Rico L, et al. Transcriptomic depression of immunological synapse as a signature of ventilator-associated pneumonia. Ann Transl Med. 2018;6(21):415.
Tawfik DM, Vachot L, Bocquet A, Venet F, Rimmelé T, Monneret G, et al. Immune profiling panel: a proof-of-concept study of a new multiplex molecular tool to assess the immune status of critically ill patients. J Infect Dis. 2020;222(Supplement_2):S84-95.
Venet F, Textoris J, Blein S, Rol ML, Bodinier M, Canard B, et al. Immune profiling demonstrates a common immune signature of delayed acquired immunodeficiency in patients with various etiologies of severe injury. Crit Care Med. 2021;37:19–22.
Friggeri A, Cazalis MA, Pachot A, Cour M, Argaud L, Allaouchiche B, et al. Decreased CX3CR1 messenger RNA expression is an independent molecular biomarker of early and late mortality in critically ill patients. Crit Care Lond Engl. 2016;20(1):204.
Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101(6):1644–55.
Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31(4):1250–6.
Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45(3):486–552.
Baxter CR. Fluid volume and electrolyte changes of the early postburn period. Clin Plast Surg. 1974;1(4):693–703.
Spahn DR, Bouillon B, Cerny V, Duranteau J, Filipescu D, Hunt BJ, et al. The European guideline on management of major bleeding and coagulopathy following trauma: fifth edition. Crit Care. 2019;23(1):98.
Le Gall JR. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA J Am Med Assoc. 1993;270(24):2957.
Vincent JL, de Mendonça A, Cantraine F, Moreno R, Takala J, Suter PM, et al. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on sepsis-related problems of the European Society of Intensive Care Medicine. Crit Care Med. 1998;26(11):1793–800.
Baker SP, O’Neill B, Haddon W, Long WB. The injury severity score: a method for describing patients with multiple injuries and evaluating emergency care. J Trauma. 1974;14(3):187–96.
Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61-111.
Demaret J, Venet F, Plassais J, Cazalis MA, Vallin H, Friggeri A, et al. Identification of CD177 as the most dysregulated parameter in a microarray study of purified neutrophils from septic shock patients. Immunol Lett. 2016;178:122–30.
Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. 2017;5(10):816–26.
Fontaine M, Planel S, Peronnet E, Turrel-Davin F, Piriou V, Pachot A, et al. S100A8/A9 mRNA induction in an ex vivo model of endotoxin tolerance: roles of IL-10 and IFNγ. PLoS ONE. 2014;9(6):e100909.
Peronnet E, Venet F, Maucort-Boulch D, Friggeri A, Cour M, Argaud L, et al. Association between mRNA expression of CD74 and IL10 and risk of ICU-acquired infections: a multicenter cohort study. Intensive Care Med. 2017;43(7):1013–20.
Peronnet E, Nguyen K, Cerrato E, Guhadasan R, Venet F, Textoris J, et al. Evaluation of mRNA biomarkers to identify risk of hospital acquired infections in children admitted to paediatric intensive care unit. PLoS ONE. 2016;11(3):e0152388.
Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4(4):259–71.
Almansa R, Ortega A, Ávila-Alonso A, Heredia-Rodríguez M, Martín S, Benavides D, et al. Quantification of immune dysregulation by next-generation polymerase chain reaction to improve sepsis diagnosis in surgical patients. Ann Surg. 2019;269(3):545–53.
Tremblay JA, Peron F, Kreitmann L, Textoris J, Brengel-Pesce K, Lukaszewicz AC, et al. A stratification strategy to predict secondary infection in critical illness-induced immune dysfunction: the REALIST score. Ann Intensive Care. 2022;12(1):76.
Landelle C, Lepape A, Voirin N, Tognet E, Venet F, Bohé J, et al. Low monocyte human leukocyte antigen-DR is independently associated with nosocomial infections after septic shock. Intensive Care Med. 2010;36(11):1859–66.
Cavaillon J, Singer M, Skirecki T. Sepsis therapies: learning from 30 years of failure of translational research to propose new leads. EMBO Mol Med. 2020. https://doi.org/10.15252/emmm.201810128.
van de Veerdonk FL, Giamarellos-Bourboulis E, Pickkers P, Derde L, Leavis H, van Crevel R, et al. A guide to immunotherapy for COVID-19. Nat Med. 2022;28(1):39–50.
Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Gordon AC, Hutton P, et al. Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am J Respir Crit Care Med. 2017;196(3):328–39.
Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun. 2018;9(1):694.
Sweeney TE, Azad TD, Donato M, Haynes WA, Perumal TM, Henao R, et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med. 2018;46(6):915–25.
Bodinier M, Peronnet E, Brengel-Pesce K, Conti F, Rimmelé T, Textoris J, et al. Monocyte trajectories endotypes are associated with worsening in septic patients. Front Immunol. 2021;12:795052.
Brakenridge SC, Starostik P, Ghita G, Midic U, Darden D, Fenner B, et al. A transcriptomic severity metric that predicts clinical outcomes in critically ill surgical sepsis patients. Crit Care Explor. 2021;3(10):e0554.
Acknowledgements
The authors would like to thank: (1) the clinical research assistants for their great help in the screening and sampling of patients as well as in the preparation of case report forms, (2) the technical staff from the Edouard Herriot Hospital-bioMérieux joint research unit for their dedicated involvement in samples processing. The authors would also like to acknowledge the important contribution and motivation of the medical and paramedical staff of the Anesthesia and Critical Care Medicine Department of the Edouard Herriot Hospital. Finally, the authors would like to thank the patients and their families for supporting this study.
Funding
The REALISM study was funded Supported, in part, by the Agence Nationale de la Recherche through a grant awarded to BIOASTER (grant number ANR-10- AIRT-03) and by bioMérieux, Sanofi, and GSK. All commercial and academic partners involved in the study agreed on the study design, collection, analysis, interpretation of data, writing of report, and decision to submit the article for publication [11]. The MIPrea was supported by funds from bioMérieux and Hospices Civils de Lyon. This project is part of Advanced Diagnostics for New Therapeutic Approaches (ADNA), a program dedicated to personalized medicine that is coordinated by Institut Mérieux and supported by the French public agency BPI France [12].
Author information
Authors and Affiliations
Contributions
All authors were involved in the analysis and interpretation of data as well as drafting the manuscript or revising it critically for important intellectual content. MB, GM, MC, AF, FC, FV, MAC, EC, EP, TR, ACL, KBP and JFL made substantial contributions to study conception and design and acquisition of data. MB, MC and JFL participated actively in statistical analysis. MB, GM, MC, AF, MAC, KBP and JFL participated actively in the interpretation of data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The IRB (Comité de Protection des Personnes Sud-Est II) approved the REALISM study (ref. 2015–42-2), and this study was registered at clinicaltrials.gov (NCT 02638779). The CIC approved the MIPrea study (IRB #5044). Informed consent was obtained from participants or their family members. The research was carried out in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
MB, MC, AF, MAC, EC, EP, KBP and JFL are bioMérieux’s employees. JFL and GM are co-inventors in patent applications covering the previous markers. bioFire—a bioMérieux company—holds patents on the technology. This does not alter the authors’ adherence to all the policies on sharing data and materials.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Table summarizing diagnostic performances of identified genes in the discovery cohort, based on OOP ROC threshold computation. Table S2. Table comparing low and high-risk patients, for previously reported biomarkers as being associated with poor outcomes in ICU. Fig. S1. ROC (Receiver Operating Characteristics) curve with AUC (area under the curve) value of the ability of the OOP-Tscore obtained at day 5–7 to distinguish between patients that will develop or not at least an ICU-acquired infection during their ICU stay
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bodinier, M., Monneret, G., Casimir, M. et al. Identification of a sub-group of critically ill patients with high risk of intensive care unit-acquired infections and poor clinical course using a transcriptomic score. Crit Care 27, 158 (2023). https://doi.org/10.1186/s13054-023-04436-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13054-023-04436-3