Abstract
Background
Ventricular septal defects (VSDs) are one of the leading causes of death due to cardiac anomalies during the first months of life. The prevalence of VSD in neonates is reported up to 4%. Despite the remarkable progress in medication, treatment and surgical procedure for VSDs, the genetic etiology of VSDs is still in infancy because of the complex genetic and environmental interactions.
Methods
Three hundred fifty subjects (200 VSD children and 150 healthy controls) were recruited from different pediatric cardiac units. Pediatric clinical and demographic data were collected. A total of six variants, rs1017 (ISL1), rs7240256 (NFATc1), rs36208048 (VEGF), variant of HEY2, rs11067075 (TBX5) and rs1801133 (MTHFR) genes were genotyped by tetra-ARMS PCR and PCR–RFLP methods.
Results
The results showed that in cases, the rs1017 (g.16138A > T) variant in the ISL1 gene has an allele frequency of 0.42 and 0.58 respectively for the T and A alleles, and 0.75 and 0.25 respectively in the controls. The frequencies of the AA, TA and TT genotypes were, 52%, 11% and 37% in cases versus 21%, 8% and 71% respectively in the controls. For the NFATc1 variant rs7240256, minor allele frequency (MAF) was 0.43 in cases while 0.23 in controls. For the variant in the VEGF gene, genotype frequencies were 0% (A), 32% (CA) and 68% (CC) in cases and 0.0%, 33% and 67% respectively in controls. The allele frequency of C and A were 0.84 and 0.16 in cases and 0.83 and 0.17 respectively in controls. The TBX5 polymorphism rs11067075 (g.51682G > T) had an allelic frequency of 0.44 and 0.56 respectively for T and G alleles in cases, versus 0.26 and 0.74 in the controls. We did not detect the presence of the HEY2 gene variant (g.126117350A > C) in our pediatric cohort. For the rs1801133 (g.14783C > T) variant in the MTHFR gene, the genotype frequencies were 25% (CC), 62% (CT) and 13% (TT) in cases, versus 88%, 10% and 2% in controls. The ISL1, NFATc1, TBX5 and MTHFR variants were found to be in association with VSD in the Pakistani pediatric cohort whilst the VEGF and HEY2 variants were completely absent in our cohort.
Conclusion
We propose that a wider programme of genetic screening of the Pakistani population for genetic markers in heart development genes would be helpful in reducing the risk of VSDs.
Similar content being viewed by others
Background
The most obvious cardiac abnormality of juveniles is ventricular septal defects (VSDs) as they encompass about 40% percent of all hereditary cardiac abnormalities in the form of an isolated defect. Ventricular septal defects are one of the leading causes of death in childhood during the first month of life [1]. The prevalence of neonatal VSD is reported up to 4% [2]. VSD can present as isolated heart disorder and as integral part of several other complex malformation including tetralogy of Fallot (TOF) and other syndromes [2]. VSD is classified into three categories on the basis of its genetic causes: VSD1 (GATA4), VSD2 (CITED2) and VSD3 (NKX2.5) [3]. Despite the remarkable progress in medication, treatment and surgical procedure for VSDs, the genetic etiology of VSDs is still in infancy due to complex genetic and environmental interaction. Certain transcription factors, termed pioneer transcription factors, play a significant role in initiation of cell programming and facilitating the cell differentiation and specification process. The ISL1 gene is one of the important pioneer factors for cardiomyocyte differentiation. ISL1 is a LIM homeodomain transcription factor expressed transiently in the second heart field [4]. At the initial stages of heart developmental, ISL1 serves as marker for progenitor cells. The human ISL1 gene is located on chromosome 5q11.1 [5]. An ISL1 gene knockout experiment in a mouse model has demonstrated the importance of this gene as knockout mice embryos died due to defects in heart development [6]. A SNP, rs1017 (NG_023040.1:g.16138A > T) in ISL1 has been reported in association with congenital heart disease in humans [7]. NFATc1 (nuclear factor of activated T cells, cytoplasmic 1) is a transcription factor that belongs to the Rel family and is required for the development of cardiac valves [8]. Gene expression in pro-valve endocardial cells is determined by NFATc1 via the activation of their heterogenic promoters [9]. The human NFATc1 gene is located on chromosome 18q23. Studies have reported genetic mutations in NFATc1 as the cause of atrio-ventricular septal defects (AVSD) [10]. A polymorphism rs7240256 (NG_029226.1:g.23449 T > C) in this gene has been reported as a risk factor of VSD. Vascular endothelial growth factor (VEGF) plays a key role in morphogenesis of the vascular system and differentiation of endothelial cells [11]. The human VEGF gene is located on chromosome 6p21.1. VEGF knockout experiments in mice have resulted in sudden death of 55% of animals. Even a single copy of the VEGF gene is not sufficient for the survival of mutant mice [12]. In humans, a polymorphism in the regulatory region of VEGF, rs36208048 (NG_008732.1:g.3877C > A) has been reported in association with VSD [13]. T-Box transcription factors (including TBX1, TBX2, TBX3, TBX5, TBX18 and TBX20) play a significant role in cardiac development such as endocardial formation and heart chamber maturation. So it is unsurprising that variants in such important transcription factors can lead to heart disorders [14]. TBX5 is primarily known for cardiac forelimb development. It also acts as transcriptional activator for the genes specifically associated with cardiomyocyte maturation. The human TBX5 gene is located on chromosome 12q24.21 [15]. Earlier studies has shown an association between the rs11067075 (NG_007373.1:g.51682G > T) in TBX5 and VSDs [16, 17].
Hairy enhancer of split related with YRPW motif protein 2 (Hey2 gene) belongs to a small family of HEY members consisting of basic helix loop helix (bHLH) transcription factors [18]. A key step in heart development is the timely and accurately addition of cardiac progenitor cells. HEY acts as regulator in the dynamics of cardiac progenitor cells [19]. The human HEY2 gene is located on chromosome 6q22.31. Genetic variants in the HEY2 gene cause heart defects [20]. A previous study reported a novel variant (NC_000006.10:g.126117350A > C) only in VSD patients [21]. MTHFR, a methylene tetrahydrofolate reductase, is an enzyme which catalyses synthesis of the initial circulatory form of folate, which in turn acts as a donor of methyl group for re-methylation of homocysteine to methionine [22]. The human MTHFR gene is located on chromosome 1p36.22. A common variant in MTHFR, rs1801133 (NG_013351.1:g.14783C > T), has been previously studied [23] and a recently published meta-analysis showed, an association of rs1801133 with the risk of heart disease [24]. Genetic variation in different ethnic groups is a confounding factor for human genetic disease research. Around the globe, genetic analysis has been performed to detect the relationship between a disease and the causative genes. In the same way, genetic research on VSD has also been conducted in different regions of the world. The aim of the present study was to determine the association of variants belonging to different genes including ISL1: rs1017 (NG_023040.1:g.16138A > T), NFATc1: rs7240256 (NG_029226.1:g.23449 T > C), VEGF: rs36208048 (NG_008732.1:g.3877C > A), TBX5: rs11067075 (NG_007373.1:g.51682G > T), HEY2: (NC_000006.10:g.126117350A > C) and MTHFR: rs1801133 (NG_013351.1:g.14783C > T), with VSD. The identification of different genetic polymorphisms in association with VSDs will aid surveillance and screening of VSD patients, facilitate genetic counselling and may help reduce the prevalence of this condition in the local region.
Methods
Ethical appsroval, written consent and recruitment of study subjects
The present study was approved by the local research ethical committee Institute of Microbiology and Molecular Genetics, University of the Punjab, Lahore Pakistan (approval # MMG1958-13/10/2020). Written informed consent was taken from the parents or guardians of patients. Two hundred samples (buccal swabs and blood) from isolated VSD patients were recruited from various pediatric cardiac wards of Lahore, Pakistan. One hundred and fifty healthy controls with no cardiac defects were included in this study. Sampling was done from 2018 to 2020.
Inclusion/exclusion criteria
The patients diagnosed with isolated ventricular septal defects were selected via ECG. The image of the heart was captured by two dimensional and Doppler echocardiography to identify the magnitude of the shunt, the size and location of the defect (the whole structure of heart). During cardiac catheterization, the blood flow rate, the Qp/Qs ratio were also measured to determine the shunt size. Echocardiograms were performed by expert pediatric cardiologists. Cases and controls with syndromic VSD, other cardiac issues and infectious diseases (seropositive for HBV/HCV/HIV) were excluded.
Data collection
Data from pediatric patients were collected on the basis of different parameters: hematological parameters like WBCs, RBCs, Hb level, platelets count, serum creatinine (SC), calcium, sodium, blood urea nitrogen (BUN), potassium, serum glutamic pyruvic transaminase, bilirubin, serum glutamic oxaloacetic transaminase, serum albumin, alkaline phosphate, and gamma GT etc., and demographic data like age, consanguineous marriage, family history, maternal Hb level during pregnancy. Use of medicine during pregnancy etc. was also recorded.
Genotyping
The pediatric samples were taken in EDTA vials and preserved at -20 °C. Genomic DNA was extracted from the human leukocytes/WBCs. The quality of extracted DNA was evaluated using an Epoch Biotek micro-plate reader (Biotek Instruments, USA) and DNA concentration was adjusted to 10 ng/uL. The tetra primer ARMS PCR technique was used for the amplification of the MTHFR, NFATc1 and TBX5 genes (Table 1). For HEY2, ISL1 and VEGF gene variants, the PCR–RFLP technique was used (Table 1). PCR reaction mixtures and conditions were optimized for each genetic marker. PCR and RFLP products were run on 2% agarose gel.
Statistical analysis
For the statistical analysis, the software SPSS version 22 was used. Genotypic frequencies of cases and controls were analysed using the chi-squared test (χ2) whereas allelic frequencies were calculated by direct counting. Genotypic and allelic frequencies were reported as percentages. Mean and standard deviation values were calculated for quantitative parameters. The study population was tested for Hardy–Weinberg equilibrium. As we analyzed 6 SNPs, the Bonferroni adjusted p-value (0.0125) was used as a threshold of significance for all analyses.
Results
Patient mean age, family history of VSD, siblings with VSD, descriptive characteristics of hematological parameters, chemical profile and blood group distribution have been explained previously [25]. For the rs1017 (g.16138A > T) variant in ISL1, the genotype frequencies were of 52% (AA), 11% (TA) and 37% (TT) in cases in comparison to 21%, 8% and 71% respectively in controls (Table 2). The frequency of the T and A alleles is 0.42 and 0.58 respectively in cases, while 0.75 and 0.25 respectively, in controls (OR: 0.242, CI: 0.158–0.37, p-value < 0.0001). The genotype frequencies deviated from Hardy–Weinberg equilibrium for the cohort (p-value: < 0.0001). The genotypic distribution in dominant and recessive models is demonstrated in Table 3 (Dominant; OR: 0.21, CI: 0.11–0.39, p-value: < 0.0001, Recessive: OR: 0.24 CI: 0.13–0.43, p-value: < 0.0001, allelic OR: 0.242, CI: 0.157–0.37, p-value: < 0.0001) (Table 3).
For the rs7240256 (g.23449 T > C) NFATc1 variant the genotype frequencies were 4% (CC), 78% (TC) and 18% (TT) in cases compared to 1.0%, 44% and 55% in controls (Table 2). The minor allele frequency (MAF) was 0.43 in cases while 0.23 in the controls (OR: 2.53, CI: 1.64–3.89, p-value < 0.0001). In the dominant model, the OR was 0.18 with CI 0.09–0.34 and p-value < 0.0001 and in the recessive model OR was 0.24 with CI: 0.03–2.23 (p-value: 0.16) (Table 3). For the control group the genotype frequencies were in Hardy–Weinberg equilibrium (p-value: 0.021). Genotyping of rs36208048 (g.3877C > A), variant in the VEGF gene showed that the percentage of the AA, CA and CC genotypes were 0%, 32% and 68% respectively in cases versus 0%, 33% and 67% in controls (Table 2). The frequency of the C and A alleles were 0.84 and 0.16 respectively in cases and 0.83 and 0.17 in controls (p-value 0.8577). For the cohort, the genotype frequencies showed deviation from Hardy–Weinberg equilibrium (p-value: 0.066). For the dominant model the OR was 1.06 with CI: 0.59–1.92 (p-value 0.84). While recessive model analysis could not be performed due to absence of homozygous genotype in cases as well as in controls (Table 3).
For the polymorphism, rs11067075 (g.51682G > T) in TBX5, the frequencies of the GG, GT and TT genotypes were 21%, 70% and 9%, respectively in cases, versus 56%, 36% and 8% in controls (Table 2). The frequency of the T and G alleles was 0.44 and 0.56 respectively in cases, while 0.26 and 0.74 in controls (OR: 2.24, CI: 1.47–3.41 (p-value 0.0002). In the dominant model, the OR was 0.21 with CI 0.11–0.38 and p-value < 0.0001 versus in the recessive model, the OR was 0.89 with CI 0.33–2.40 and p-value 0.82 (Table 3). For the control group, the genotype frequencies were in Hardy–Weinberg equilibrium (p-value: 0.60). For the variant in HEY2: (g.126117350A > C), we did not detect the homozygous or the heterozygous genotypes. Therefore, dominant and recessive model analysis could not be performed in the cases or in controls. Dominant homozygous frequency was 1.0, both in cases and controls. The genotyping of rs1801133 (g.14783C > T) variant in MTHFR showed that the frequencies of the CC, CT and TT genotypes were 25%, 62% and 13%, respectively in cases, while 88%, 10% and 2% in controls (Table 2). The allelic frequency of T and C are 0.56 and 0.44 respectively, in cases, while 0.93 and 0.07 in controls (OR: 10.46, CI: 5.68–19.26, p-value (< 0.0001). In the dominant model, the OR was 0.04 with CI 0.02–0.10 (p-value < 0.0001) while in the recessive model, the OR was 0.14 with CI 0.33–0.63 and p-value 0.002 (Table 3). For the control group, the genotypic frequencies were in Hardy–Weinberg equilibrium (p-value: 0.65).
Discussion
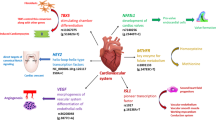
The healthcare burden of congenital heart disease is gradually increasing worldwide and more specifically in the low- and middle- income countries like Pakistan, where the cost of treatment is unaffordable for most families. Generally, the Pakistani population shows remarkable variations in the investigation of genetic diseases due to high rates of consanguineous marriages, a fact that makes this population attractive to geneticists. Advances in the field of genetics have led to the discovery of novel genetic markers for complex as well as for monogenic diseases [1]. Ventricular septal defect is the commonest congenital heart defect with 1 in 500 live births. Risk factors for VSD include genetic variations, family history, ethnicity and certain environmental factors. According to the data reported in previous studies, genes involved in the circulatory system and cardiac development such as, GATA4, NKX2.5, TBX5, VEGF, MTRR, MTHFR, ISL1, NFATC1 and CITED2 play crucial roles in the etiology of VSD [26]. Due to unavailability of pre- and post-natal biomarkers in clinical practice it is crucial to identify genetic risk or disease markers for VSDs. All genes (ISL1, NFATc1, VEGF, HEY2, TBX5 and MTHFR) selected in this study are important structurally as well as functionally in cardiovascular system (Fig. 1).
The ISL1 LIM-homeodomain transcription factor substantially contributes to embryonic heart development. It is one of the genes responsible for ventricle development [26]. Based on a previous study on a Chinese cohort, we selected rs1017 polymorphism as potential biomarker for prediction of VSD [27]. A study of meta-analysis suggested this polymorphism as a risk factor for heart disease [5]. In contrast to the Chinese population, no association was detected in a White population [28]. Surprisingly, in the current study, results showed the minor allele as the protective allele for VSDs in the Pakistani population. This was confirmed by genetic model analysis. Valvular and septal development is greatly affected by NFATc1. An earlier study reported significant association of variant homozygous genotype of rs7240256 with VSDs [29]. Our results are in consensus with a previous study where rs7240256 showed association with complications of VSDs [30]. Higher proportion of the risk genotype is an indicator of the effect of this polymorphism on valvular and septal development. VEGF is a signaling protein involved in angiogenesis. Any dysregulation in the function and structure of VEGF, can play a critical role in the pathogenesis of VSDs. Circulatory vasculature is under direct influence of growth factors like VEGF, any deviation from the normal function of this growth factor can lead to serious consequences [31]. From our findings, selected SNP rs36208048 of VEGF did not show statistical difference in allele frequencies between cases and controls despite this variant being located in the promoter region of gene which is an essential part for the normal function of the growth factor [13].
TBX5 transcription factors maintained their expression level throughout heart development and their persistent expression has also been observed in ventricular maturation [15, 32]. Our results are in concordance with a previous study, where variant rs11067075 in TBX5 was associated with VSD in pediatric patients [16]. In our study population, we observed different minor allele frequencies in cases and control with the risk allele playing a role in the association between variant and outcome. Dominant model analysis also supported these results. For the cardiac developmental pathways, HEY2 transcription factor plays an important role along with other factors like NKX2.5, TBX5 and GATA4 [33]. A study on a German population reported a novel polymorphism in the binding domain of HEY c.293A > C (p.Asp98Ala) (g.126117350A > C) [21]. Due to low sample size, sample bias and ethnic difference we did not find any significant difference in allelic and genotypic frequency in current cohort. However, this variation has been seen rarely, possibly due to its functionally important position in the HEY2 transcription factor. DNA, RNA and proteins need one carbon (methyl group) for methylation and re-methylation, usually catalysed by MTHFR. MTHFR also catalyses the conversion of homocysteine into methionine. Based on data published previously, the T allele of rs1801133 (g.14783C > T), is a risk factor for heart disorders [34,35,36]. Different ethnic groups including Egyptians, Tamilians, Iranian and Chinese have been reported to carry the T allele in association with congenital heart disease [22, 37,38,39,40]. Similarly, we obtained results in our cohort showing that the risk allele has a higher frequency in cases compared to controls. This suggests a significant association of the T allele with VSDs in the local ethnic groups.
Conclusion
The Pakistani population is unique, in regard of its social, cultural, religious practices; it also has been reported to have a high percentage of consanguinity. This makes genetic research and in particular the study of complex genetic disorders including VSDs extremely challenging, but potentially rewarding. Although samples can be recruited from the entire country, their total number is limited due to lack of resources. This is the first attempt to determine the possible association of variants in the ISL1, NFATc1, VEGF, HEY2, TBX5 and MTHFR genes with VSDs in a Pakistani cohort.
Availability of data and materials
All data is available with the corresponding author and can be accessed on request.
Abbreviations
- VSD:
-
Ventricular septal defects
- VEGF:
-
Vascular endothelial growth factor
- TBOX:
-
T-Box transcription
- ISL1:
-
ISLET1
- MTHFR:
-
Methylene tetrahydrofolate reductase
- SPSS:
-
Statistical Package for Social Sciences
References
Sarwar S, Ehsan F, Tahir A, Jamil M, Shahid SU, Khan A, et al. First report of polymorphisms in MTRR, GATA4, VEGF, and ISL1 genes in Pakistani children with isolated ventricular septal defects (VSD). Ital J Pediatr. 2021;47(1):1–6.
Penny DJ, Vick GW III. Ventricular septal defect. Lancet. 2011;377(9771):1103–12.
Sarwar S, Hasnain S. Classification of ventricular septal defects (VSDS) on the basis of genetics. Pakistan Heart J. 2020;53(4):285–91.
Gao R, Liang X, Cheedipudi S, Cordero J, Jiang X, Zhang Q, et al. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res. 2019;29(6):486–501.
Ding Z, Yang W, Yi K, Ding Y, Zhou D, Xie X, et al. Correlations between ISL1 rs1017 polymorphism and congenital heart disease risk: a PRISMA-compliant meta-analysis. Medicine. 2020;99(2):e18715.
Ma L, Wang J, Li L, Qiao Q, Di R-M, Li X-M, et al. ISL1 loss-of-function mutation contributes to congenital heart defects. Heart Vessels. 2019;34(4):658–68.
Luo Z, Sun H, Yang Z, Ma Y, Gu Y, He Y, et al. Genetic variations of ISL1 associated with human congenital heart disease in Chinese Han people. Genet Mol Res. 2014;13(1):1329–38.
Wu B, Baldwin HS, Zhou B. Nfatc1 directs the endocardial progenitor cells to make heart valve primordium. Trends Cardiovasc Med. 2013;23(8):294–300.
Gunawan F, Gentile A, Gauvrit S, Stainier DY, Bensimon-Brito A. Nfatc1 promotes interstitial cell formation during cardiac valve development in zebrafish. Circ Res. 2020;126(8):968–84.
Ferese R, Bonetti M, Consoli F, Guida V, Sarkozy A, Lepri FR, et al. Heterozygous missense mutations in NFATC1 are associated with atrioventricular septal defect. Hum Mutat. 2018;39(10):1428–41.
Luo B, Wu Y, Liu S-l, Li X-y, Zhu H-r, Zhang L, et al. Vagus nerve stimulation optimized cardiomyocyte phenotype, sarcomere organization and energy metabolism in infarcted heart through FoxO3A-VEGF signaling. Cell Death Dis. 2020;11(11):1–16.
Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, et al. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130(4):691–703.
Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12(8):1232–5.
Khatami M, Heidari MM, Kazeminasab F, Bidaki RZ. Identification of a novel non-sense mutation in TBX5 gene in pediatric patients with congenital heart defects. J Cardiovasc Thorac Res. 2018;10(1):41.
Steimle J, Moskowitz I. TBX5: a key regulator of heart development. Curr Top Dev Biol. 2017;122:195–221.
Liu C-x, Shen A-d, Li X-f, Jiao W-w, Song B, Feng Y, et al. Association of TBX5 gene polymorphism with ventricular septal defect in the Chinese Han population. Chin Med J. 2009;122(1):30–4.
Bellmann K, Perrot A, Rickert-Sperling S. Human genetics of ventricular septal defect. Congenital Heart Diseases: The Broken Heart. Springer; 2016. p. 307–28. ISBN: 978-3-7091-1882-5.
Weber D, Wiese C, Gessler M. Hey bHLH transcription factors. Curr Top Dev Biol. 2014;110:285–315.
Gibb N, Lazic S, Yuan X, Deshwar AR, Leslie M, Wilson MD, et al. Hey2 regulates the size of the cardiac progenitor pool during vertebrate heart development. Development. 2018;145(22):dev167510.
van Walree ES, Dombrowsky G, Jansen IE, Mirkov MU, Zwart R, Ilgun A, et al. Germline variants in HEY2 functional domains lead to congenital heart defects and thoracic aortic aneurysms. Genet Med. 2021;23(1):103–10.
Reamon-Buettner SM, Borlak J. HEY2 mutations in malformed hearts. Human Mutat. 2006;27(1):118-.
Noori N, Miri-Moghaddam E, Dejkam A, Garmie Y, Bazi A. Are polymorphisms in MTRR A66G and MTHFR C677T genes associated with congenital heart diseases in Iranian population? Caspian J Intern Med. 2017;8(2):83.
Zhang R, Huo C, Wang X, Dang B, Mu Y, Wang Y. Two common MTHFR gene polymorphisms (C677T and A1298C) and fetal congenital heart disease risk: an updated meta-analysis with trial sequential analysis. Cell Physiol Biochem. 2018;45(6):2483–96.
Liu P-F, Ding B, Zhang J-Y, Mei X-F, Li F, Wu P, et al. Association between MTHFR C677T polymorphism and congenital heart disease a PRISMA-compliant meta-analysis. Int Heart J. 2020;61(3):19–389.
Sarwar S, Ehsan F, Tahir A, Jamil M, Shahid SU, Hasnain S, et al. Hematological and demographic profile of Pakistani children with isolated ventricular septal defects (VSDs). Egypt J Med Human Gen. 2020;21(1):1–8.
Radhakrishna U, Albayrak S, Zafra R, Baraa A, Vishweswaraiah S, Veerappa AM, et al. Placental epigenetics for evaluation of fetal congenital heart defects: Ventricular Septal Defect (VSD). PLoS ONE. 2019;14(3):e0200229.
Lang J, Tian W, Sun X. Association between ISL1 variants and susceptibility to ventricular septal defect in a Chinese cohort. Mol Diagn Ther. 2013;17(2):101–6.
Cresci M, Vecoli C, Foffa I, Pulignani S, Ait-Ali L, Andreassi MG. Lack of association of the 3′-UTR polymorphism (rs1017) in the ISL1 gene and risk of congenital heart disease in the white population. Pediatr Cardiol. 2013;34(4):938–41.
Gu H, Gong J, Qiu W, Cao H, Xu J, Chen S, et al. Association of a tandem repeat polymorphism in NFATc1 with increased risk of perimembranous ventricular septal defect in a Chinese population. Biochem Genet. 2011;49(9–10):592.
Lei S, Li Z-z, Shen A-d, Hui L, Song B, Jian G, et al. Association ofNFATc1gene polymorphism with ventricular septal defect in the Chinese Han population. Chin Med J. 2013;126(1):78–81.
Liu F, Hidru TH, Gao R, Lin Y, Liu Y, Fang F, et al. Cancer patients with potential eligibility for vascular endothelial growth factor antagonists use have an increased risk for cardiovascular diseases comorbidities. J Hypertens. 2020;38(3):426.
Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol. 2000;223(1):169–80.
Anderson DJ, Kaplan DI, Bell KM, Koutsis K, Haynes JM, Mills RJ, et al. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat Commun. 2018;9(1):1–13.
Luo Z, Lu Z, Muhammad I, Chen Y, Chen Q, Zhang J, et al. Associations of the MTHFR rs1801133 polymorphism with coronary artery disease and lipid levels: a systematic review and updated meta-analysis. Lipids Health Dis. 2018;17(1):1–15.
Raina JK, Sharma M, Panjaliya RK, Dogra V, Bakaya A, Kumar P. Association of ESR1 (rs2234693 and rs9340799), CETP (rs708272), MTHFR (rs1801133 and rs2274976) and MS (rs185087) polymorphisms with Coronary Artery Disease (CAD). BMC Cardiovasc Disord. 2020;20(1):1–13.
Huang J, Mei J, Jiang L, Jiang Z, Liu H, Ding F. MTHFR rs1801133 C> T polymorphism is associated with an increased risk of tetralogy of Fallot. Biomed Rep. 2014;2(2):172–6.
Shi H, Yang S, Lin N, Huang P, Yu R, Chen M, et al. Study on maternal SNPs of MTHFR Gene and HCY level related to congenital heart diseases. Pediatr Cardiol. 2021;42(1):42–6.
Shi H, Yang S, Liu Y, Huang P, Lin N, Sun X, et al. Study on environmental causes and SNPs of MTHFR, MS and CBS genes related to congenital heart disease. PLoS ONE. 2015;10(6):e0128646.
Zidan HE, Rezk NA, Mohammed D. MTHFR C677T and A1298C gene polymorphisms and their relation to homocysteine level in Egyptian children with congenital heart diseases. Gene. 2013;529(1):119–24.
Angeline T, Jeyaraj N, Granito S, Tsongalis GJ. Prevalence of MTHFR gene polymorphisms (C677T and A1298C) among Tamilians. Exp Mol Pathol. 2004;77(2):85–8.
Acknowledgements
Professor Ann Walker, Dr. Petros Syrris, Institute of Cardiovascular Science, University College London are acknowledged for their kind help in manuscript writing and technical assistance.
Funding
This work was supported by the Higher Education Commission of Pakistan, Grant # [518–111767-2BS5-019].
Author information
Authors and Affiliations
Contributions
Shabana and SHconceived the study concept, AT, ZL, SN, SM, and RSS carried out bench work, SS, SUS and Shabana analyzed the results, and drafted the manuscript, Shabana critically reviewed the manuscript and supervised the study. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the local research ethical committee, Institute of Microbiology and Molecular Genetics, University of the Punjab, Lahore Pakistan (approval # MMG1958-13/10/2020). Written informed consent was taken from the parents or guardians of patients. All methods were carried out in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Consent for publication
There is no conflict of interests regarding the publication of this article.
Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sarwar, S., Shabana, Tahir, A. et al. Study of variants associated with ventricular septal defects (VSDs) highlights the unique genetic structure of the Pakistani population. Ital J Pediatr 48, 124 (2022). https://doi.org/10.1186/s13052-022-01323-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13052-022-01323-5