Abstract
Objectives
The aim of the study is to explore the relationship between CCDC69 expression and resistance of ovarian cancer cells to cisplatin and reveal the underlying mechanism.
Methods
One hundred thirty five ovarian cancer patients with intact chemo-response information from The Cancer Genome Atlas (TCGA) database were included and analyzed. Stable CCDC69 overexpressing 293 and ovarian cancer A2780 cell lines were established and subjected to examine cell apoptosis and cell cycle distribution using CCK-8 assay and flow cytometry. Cell cycle and apoptosis pathway were evaluated by immunoblots. Stability of p14ARF/MDM2/p53 pathway related proteins were determined by half-life analysis and ubiquitination experiments.
Results
We found that CCDC69 expression was significantly higher in chemo-sensitive groups compared with chemo-resistant groups from TCGA database. High CCDC69 expression was associated longer survival. CCDC69 overexpressing 293 and A2780 cells with wildtype p53 and contributes to cisplatin sensitivity following treatment with cisplatin. We further found over-expression of CCDC69 activated p14ARF/MDM2/p53 pathway. Importantly, we also demonstrated that CCDC69 expression extended p53 and p14ARF protein half-life and shortened MDM2 protein half-life. Ubiquitination assay revealing a decrease in p14 ubiquitination in CCDC69 over-expression cells comparing to cells expressing empty vector.
Conclusions
It is tempting to conclude that targeting CCDC69 may play a role in cisplatin resistance.
Similar content being viewed by others
Introduction
Epithelial ovarian cancer (EOC) is the second most common and accounts for greatest number of death from gynecologic malignancies in the world wide [1, 2]. High-grade serous ovarian cancer (HGSC) is an aggressive subtypes and often associated with relatively unfavorable clinical outcomes [3]. Majority of advanced ovarian cancer patients initially responsive to chemotherapy; however, became increasingly ineffective and eventually results in treatment failure [4]. Thus, exploring and understanding the mechanisms responsible for chemo-resistance in ovarian cancer may improve the therapeutic outcomes.
Cisplatin has been the effective and widely used drugs against various human cancers, including ovarian cancer [5]. In response to cisplatin, p53, a key regulator of cell cycle checkpoints, activates a number of genes regulating cells cycle and/or apoptosis [6]. As transcriptional target of p53, p21 plays an important role in the maintenance of and G1 and G2 checkpoints following DNA damage such as cisplatin exposure [6,7,8]. In addition, p21 can regulate cell cycle progression dependent or independent of p53 following treatment with cisplatin [9, 10]. Previous reports have shown that abrogation of G2/M cell cycle checkpoint is related to enhancement of cisplatin-induced cytotoxicity [11, 12]. Expression of the p53 protein is posttranscriptionally regulated by MDM2. The MDM2 binds to p53 and blocking its activation domain [13], thus promoting degradation of p53 via ubiquitin-proteasome pathway [14, 15]. Under the DNA damage pressure, the p53 phosphorylation attenuates MDM2 binding ability and then leads to p53 accumulation [16]. Until recent years, investigators have identified another important regulator in this p53/MDM2 pathway, the cyclin-dependent kinase inhibitor p14ARF (termed p19ARF in the mouse) [17]. The p14ARF protein is an alternative reading frame protein product of the CDKN2A/INK4A locus [18]. The other one is p16INK4A protein [17]. It has been shown that the p14ARF binds to the p53/MDM2 complex and inhibits MDM2-mediated degradation of p53 [19]. p14ARF can inhibit cell cycle progression in both G1 and G2/M phases and/or cell apoptosis in a p53-dependent and independent manner [20, 21]. Furthermore, apoptosis stimulated by p14ARF apoptosis is enhanced by loss of p21 mediated cell cycle checkpoint control [22].
Coiled-coil domain-containing (CCDC) proteins have variety of functions in regulating cell cycle progression and mediating apoptosis following DNA damage in eukaryotic cells [23, 24]. In recent years, several CCDC proteins have been widely studied as tumor suppressors in several types of cancer, such as breast and prostate cancers non-small cell lung cancer [25,26,27]. However, the studies on the role of coiled-coil domain-containing 69 (CCDC69) in ovarian cancer were limited. As one of these family members, CCDC69 locates on 5q33.1 and is responsible for mitotic spindle and DNA replication [28]. In the present study, we found that CCDC69 expression is significantly higher in chemo-sensitive groups compared with chemo-resistant groups from The Cancer Genome Atlas (TCGA) database. We also found women with high CCDC69 expression was related to longer survival. To further elucidate the role of CCDC69, we then stably expressed CCDC69 in 293 cells and human ovarian cancer cell lines A2780 with functional p53. Our data showed that expression of CCDC69 abrogates G2/M arrest followed by apoptosis in these p53 wildtype cells. Importantly, we also demonstrated that CCDC69 expression extended p53 and p14ARF protein half-life and shortened MDM2 protein half-life due to deubiquitination of p14ARF.
Materials and methods
Chemo-response and survival analysis using public datasets
TCGA clinical and CCDC69 expression mRNA data were retrieved from published The Cancer Genome Atlas (TCGA) through the Computational Biology Center Portal (cBio): http://www.cbioportal.org/.The cgdsr extension package was used to execute the retrieval.
Cell lines
Human ovarian cancer cell line A2780 was purchased from Sigma-Aldrich and routinely maintained in RPMI 1640 (Invitrogen) supplemented with heat-inactivated 10% (v/v) fetal bovine serum (Invitrigen) and 1% antibiotic-antimycotic. The human embryonic kidney cells 293 was purchased from ATCC (Bethesda, MD, USA). 293 cells were cultured in high-glucose DMEM (Invitrogen) containing 10% fetal bovine serum (FBS) (Invitrogen) and 100 U/ml of penicillin G/streptomycin. These mammalian cells were cultured in an incubator with an atmosphere of 37 °C in 5% CO2. Cell culture medium was refreshed every two days.
Mutation analysis of p53
Polymerase chain reaction (PCR) amplification and direct sequencing were used to screen for DNA variations in the coding and the 5′ and 3′ flanking regions of p53. Primers for exons 5–8 are modified from Martin et al. [29]. Primers used for sequencing not shown Additional file 1: Table S1. All purified PCR products were sequenced for variations using an ABI PRISM® Big Dye™ Terminator version 3.1 Cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3100 automated sequencer (Applied Biosystems).
Viral packaging and lentiviral transduction
For the production of lentiviral particles, 293 (1 × 106) were transfected with CCDC69 lentiviral construct (pLenti-CCDC69-GFP) or control plasmid (pLenti-C-GFP, Origene, Bothell, WA, USA) together with pRSV-Rev and psPAX2 packaging vectors using Lipofectamine 2000 transfection reagent (Invitrogen). To generation stable cell lines, viral supernatant were harvested and filter through a 0.45 μm filter to remove cellular debris. The viral supernatant and 4.5 mL of fresh growth medium were added to plates and incubate the cells at 37 °C with 5% CO2 for 4 h. And then removed the transduction medium and added 10 mL of fresh growth medium. Incubated the cells for three more days in the presence of puromycin, and resistant colonies were selected for subsequent experiments.
Cell viability assay
Cell viability was measured by MTT assay using the Cell Counting Kit-8 (CCK-8, Sigma-Aldrich) following the manufacturer’s instructions. The absorbance at 450 nm was measured in the microplate reader. The percentage of cell survival at each dose of cisplatin = Mean of A450 (drug-treated cells)/Mean of A450 (untreated cells). IC50 values were calculated using GraphPad Prism Software Version 5 (GraphPad Software Inc., CA, USA) and plotted in dose response curves.
Flow cytometry analysis of cell apoptosis using Annexin V-FITC/PI staining
The experiments was performed according to the manual of Alexa Fluor® 488 Annexin V/Dead Cell Apoptosis Kit (Invitrogen) by flow cytometry. About 1 × 106 cells were collected, washed with ice-cold PBS, and resuspended in binding buffer containing suitable amount of Annexin V-FITC. After 15 min of incubation in the dark at room temperature, the buffer was removed by centrifugation. The cells were then resuspended in reaction buffer containing propidium iodide (PI). Apoptosis was immediately detected by flow cytometry.
Cell cycle assay
After the indicated treatments, cells were washed with cold PBS and harvested by centrifugation. Then, cells were re-suspended in 70% (v/v) cold ethanol and stored at − 20 °C overnight. After 30-min incubation with propidium iodide (PI) solution in the dark, cell cycle distribution was analyzed by Cytomics FC 500 (Beckman Coulter) flow cytometer. Results were calculated and visualized by Flow Jo software (version 10.0).
Antibodies
Antibodies against CCDC69, p21 and p14ARF were from Abcam (ab106692, ab109199 and ab3642). p53 Antibody (DO-1) and MDM2 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HRP-conjugated anti-rabbit and anti-chicken IgG antibody were used as secondary antibody from Abcam (ab191866 and ab6877). GAPDH was used as an internal control for normalization of protein quantity.
Real-time PCR
RNA was extracted with the RNeasy Mini Kit (Qiagen). cDNA was generated with PrimeScript cDNA Synthesis Kit (Takara). P21 mRNA expression levels were evaluated by real-time PCR using the ABI PRISM 7500 sequence detection system (Applied Biosystems, Foster City, CA). TaqMan™ Gene expression assays [cyclin dependent kinase inhibitor 1A (CDKN1A)/p21 (Hs00355782_m1)] were from Applied Biosystems. Quantification was based on a comparative ΔCt method using glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs03929097_g1) as endogenous control. Each real-time RT-PCR was done in triplicate according to the manufacturer’s instructions.
Half-life analysis
CCDC69 overexpressing 293 or A2780 cells and controls were incubated with cycloheximide (100 μg/mL; Sigma) to inhibit further protein synthesis. Following incubation for 30, 60, or 120 min, cells were harvested. Western blot was done as described above. The relative p53, MDM2, and p14ARF levels were quantified by densitometry analysis using the ImageJ 1.410 image processing software.
Immunoprecipitation
For immunoprecipitation/immunoblotting to detect ubiquitinated forms of p14ARF, cells treated with or without cisplatin in the presence of MG132 (20 μM). After incubations for 24 h, cells were lysed in RIPA buffer and protease and phosphatase inhibitor cocktail (Thermo Scientific). Protein lysates were precleared with protein G agarose beads (Millipore) for 1 h and then incubated with G-protein beads bound to p14ARF rabbit polyclonal antibody (Abcam) for 2 h at 4 °C. Beads were washed 3 times in RIPA buffer. Protein was eluted from beads with 2 x SDS-β-mercaptoethanol sample buffer, boiled for 8 min and then loaded on polyacrylamide gels for SDS–PAGE as described above. Blots were blocked in BSA 5% (for phosphor antibodies) or non-fat dry milk 5% in TBS–Tween 0.1% for 1 h and then incubated with primary antibody. Samples were subjected to SDS-PAGE, followed by immunoblotting with indicated antibodies.
Data statistical analysis
Data from at least three independent experiments were expressed as mean ± standard deviation (SD). The differences between two groups were analyzed using the Student’s t test. All analysis was performed using GraphPad Prism Software Version 5 (GraphPad Software Inc., La Jolla, CA, USA) at a two-sided 5% significance level.
Results
CCDC69 differentially expressed in chemo-sensitive and chemo-resistant groups
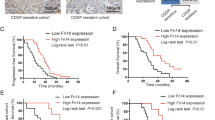
A total of 135 patients diagnosed with high-grade serous ovarian carcinoma with intact chemo-response information from The Cancer Genome Atlas (TCGA) provisional dataset were recruited into the study. The analysis revealed that CCDC69 expression is significantly higher in chemo-sensitive groups compared with chemo-resistant groups (p = 0.0080) (Fig. 1a). The relationship between patient demographic variables and ovarian cancer chemo-response was analyzed (Table 1). Moreover, survival analysis using ovarian cancer microarray datasets (GSE9891 and GSE17260) indicated a significant association between high CCDC69 expression and better patient survival in the PrognoScan database [30,31,32] (Fig. 1 b and c, p = 0.033 and p = 0.044, respectively). These data indicate that CCDC69 expression confers to chemo-sensitive and plays a protective role in cancer cell survival.
Higher CCDC69 gene expression correlates with increased survival of ovarian cancer patients. a, dot plot for expression of CCDC69 in chemo-sensitive groups and chemo-resistant group using TCGA database. **p < 0.01 versus resistant group (Student’s t-test). b, survival analysis for high-grade serous ovarian carcinoma patient using Gene Expression Omnibus (GEO) database
Overexpression of CCDC69 sensitizes 293 and A2780 cells to cisplatin
To understand the role of CCDC69 for cisplatin sensitivity to cells, A2780 and 293 cells were lentiviral transduced with a GFP tagged CCDC69 expression vector or with GFP as a negative control and cultured with puromycin (3 μg/ml) for 14 days. Exogenously expressed CCDC69 was detected by immunofluorescence staining (Data not shown). Immunoblot analysis confirmed that a higher CCDC69 expression in the CCDC69 overexpressing cells compared to those expressing an empty vector (Fig. 2c).
CCDC69 confers chemo-sensitivity in 293 and A2780 cells. a. Sensitization of cells to cisplatin after CCDC69 overexpression as revealed by the CCK-8 cytotoxicity assay. b. Apoptosis was analyzed by flow cytometry after annexin V and propidium iodide staining. Total apoptosis is the sum of the percentage of annexin V only and annexin V/propidium iodide stained cells. c. immunoblot analysis of CCDC69 and cleaved PARP in 293 cells after stable CCDC69 overexpression and treatments with cisplatin for 48 h. Loading control, GAPDH
CCDC69 overexpressing 293 and A2780 cells showed an increase in cisplatin sensitivity compared to the cells expressing GFP (Fig. 2a). Moreover, an increased annexin V percentages of positive cells and higher levels of cleaved PARP were found in CCDC69 overexpressing 293 and A2780 cells compared to those expressing an empty vector in the presence of cisplatin treatment (Fig. 2b and c).
As a key molecule regulating apoptosis, we found that p53 protein levels were profoundly increased in CCDC69 overexpressing 293 cells compared to cells expressing empty vector treatment with or without cisplatin (Fig. 2c). Besides, DNA direct sequencing data showed no p53 mutations in A2780 and 293 cells. Collectively, these data indicate that CCDC69 plays an important role in enhancing cells to cisplatin-induced cell death.
Downregulation of p21 in CCDC69 overexpressing 293 cells during cisplatin treatment arrest G2 arrest
As one of the downstream target of p53, we next assessed the expression of p21 by Western blot. The data showed that p21 was marked decreased in CCDC69 overexpressing 293 cells than cells expressing empty vector (Fig. 2c). We further determine the cell cycle phase distribution in CCDC69 overexpressing 293 cells and cells expressing empty vector using flow cytometry. We found that CCDC69 overexpressing 293 cells had significant lower percentages of G2/M phase (Fig. 3). Consistent with apoptotic experiments, we found obvious accumulation of CCDC69 overexpressing cells at sub-G1 (Fig. 3a), which is a clear indicator of apoptosis.
CCDC69 overexpressing cells showed abrogated G2/M arrest after cisplatin treatment. 293 wildtype and 293 CCDC69 overexpressing cells were treated with 50 μM cisplatin for 48 h, and then cell cycle was analyzed by flow cytometry. Data represent the mean and the standard deviation from three independent experiments. *p < 0.05 and **p < 0.01 versus cisplatin-treated 293 wildtype cells (Student’s t-test)
It has been reported that stabilized or accumulated p53 without induction of p21 protein level, suggesting that the induced p53 was transcriptionally inactive [33,34,35]. We therefore investigated the expression of p21 mRNA in both CCDC69 overexpressing 293 cells and cells expressing empty vector. An increase in p21 mRNA expression was seen at 8 h and 24 h following treatment with cisplatin (Fig. 3b). Collectively, these results indicate that abrogated cell cycle phase may due to p53-independent downregulation of p21 following treatment with cisplatin.
CCDC69 regulates p53/MDM2/p14ARF signaling pathway
To explain the accumulation of p53, the p14ARF/MDM2/p53 pathway related proteins were evaluated. We found marked reduction in the expression of p14ARF in CCDC69 overexpressing 293 and A2780 cells. These results indicate that CCDC69 enhances the accumulation of p53 may result from inhibition of p14ARF after overexpression of CCDC69.
Therefore, to further determine whether p14 promoted MDM2 degradation is involved in the accumulation of p53 in the presence of CCDC69, we assessed the expression of p14, p53 and MDM2 after treatment with the inhibitor of protein biosynthesis CHX (cycloheximide) (100 μg/ml) for various lengths of time (Fig. 4a). As it was shown in Fig. 4a, the data showed that CCDC69 overexpression significantly extended the half-life of p53 and p14ARF, on the contrary, shortened the half-life of MDM2 (Fig. 4a), suggesting that CCDC69 promotes the accumulation of p53 through activating p14ARF while inactivating MDM2 signaling to sustain p53 and p14ARF expression during cisplatin exposure. Furthermore, ubiquitination assay revealing a decrease in p14 ubiquitination in CCDC69 overexpression cells comparing to cells expressing empty vector (Fig. 4b).
CCDC69 regulates p53, MDM2, and p14ARF stability. a. 293 wildtype and 293 CCDC69 overexpressing cells were treated with 100 μg/mL cycloheximide (CHX) for the indicated periods, followed by Western blot using antibody against p53, MDM2, p14ARF, or CCDC69, with GAPDH as the loading control. The relative p53, MDM2, and p14ARF levels were quantified by densitometry analysis (right). b. Ubiquitylation assay revealing a gradual increase in p14ARF ubiquitylation from CCDC69 overexpression 293 cells. IP, immunoprecipitation; IB, immunoblot
Discussion
The p14ARF/MDM2/p53 pathway has been well established signaling axis in determining cancer cells apoptosis/quiescence or senescence [36]. It is worth noting that p14 could also induce p53-independent apoptosis [20, 21]. Furthermore, p14 triggered apoptosis is enhanced by loss of p21 mediated cell cycle checkpoint by p53-mediated mitochondrial apoptotic cascade [22, 37]. In our study, immunoblots analyses in p53 wildtype cells indirectly suggested that p14 is dispensable for accelerating cell death after induction of CCDC69 following treatment with cisplatin. Most intriguingly, it was reported recently that p53 aggregation could sensitize cancer cells to platinum treatment [38]. The most likely explanation is that these controversial findings may come from different populations of cancer cells properties. Therefore, the exact nature of the molecular mechanisms involved in these processes need for further investigation in the future.
In our study, we demonstrated that CCDC69 expressing cells abrogated G2 arrest in the reduction expression of p21. Considering that p14 could induce cell cycle arrest in both G1 and G2 phases in a p53-dependent and independent manner [39,40,41]. One of the explanation is that p21 might be the major cause for G2 cell cycle arrest in response to CCDC69 induction following treatments with cisplatin. Moreover, we showed that reduction of p21 expression in an entirely p53-independent fashion, as the p21 mRNA was found to be transcriptionally inactive. We excluded genetic mutation for p53 inactivation and learnt that many tumors without p53 mutation still harbor a transcriptionally inactive form of p53 [42, 43]. These findings raise the question of the importance of cell cycle variations in cancer cells.
Numerous studies have demonstrated that the tumor suppressor protein p14ARF can stabilize and activate p53 by inhibiting the E3 ligase activity and nuclear export of MDM2, which is followed by formation of the p14ARF-MDM2-p53 ternary complex [44,45,46]. Based on the extended half-life and decreased ubiquitination level of p14ARF in the CCDC69 overexpressing cells, we concluded that CCDC69 was able to activate p14ARF by increasing stability of the p14ARF protein, which, in turn, promotes activation p53 pathway. Based on above findings, CCDC69 mediates stability of p14ARF and regulates cell cycle and apoptosis response to chemotherapy. Therefore, CCDC69 may be a promising target for therapeutic application in cancer. Although p53 mutations are most frequently occurred in cancer [47, 48], researchers had defined an activity of p14ARF in sensitizing p53 mutated osteosarcoma cells to cisplatin [49]. However, we did not assess the ubiquitination status of p53 and MDM2. Moreover, the physical interactions between p14 and CCDC69 should be further investigated in the future.
In summary, we present data showing that the expression of CCDC69 in chemosensitive groups was significant higher than chemoresistance groups. In addition, the high expression of CCDC69 was associated with better survival based on publicly available databases. Furthermore, CCDC69 could activate the p14ARF/MDM2/p53 signaling pathway, resulting in cancer cell apoptosis. Thus, our study provide knowledge about the role of CCDC69 in chemoresistance of ovarian cancer and provide us further clue develop new therapeutic strategy for ovarian cancer patients.
Conclusion
In conclude, CCDC69 could be play important roles in regulating chemoresistance through activation of p14ARF/MDM2/p53 signaling pathway.
Abbreviations
- BSA:
-
Bovine serum albumin
- cBio:
-
computational biology center portal
- CCDC:
-
Coiled coil domain containing
- CCDC69:
-
Coiled coil domain containing 69
- CCK-8:
-
Cell Counting Kit-8
- CHX:
-
Cycloheximide
- DMEM:
-
Dulbecco modified eagle medium
- EOC:
-
Epithelial ovarian cancer
- FBS:
-
Fetal bovine serum
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- HGSC:
-
High-grade serous ovarian cancer
- IB:
-
Immunoblotting
- IP:
-
Immunoprecipitation
- MDM2:
-
Mouse double minute 2 homolog
- MTT:
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PCR:
-
Polymerase chain reaction
- PI:
-
Propidium iodide
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCGA:
-
The Cancer Genome Atlas
References
Meserve EE, Brouwer J, Crum CP. Serous tubal intraepithelial neoplasia: the concept and its application. Mod Pathol. 2017;30(5):710–21.
Chan JL, Wang ET. Oncofertility for women with gynecologic malignancies. Gynecol Oncol. 2017;144(3):631–6.
Tanenbaum LM, Mantzavinou A, Subramanyam KS, Del Carmen MG, Cima MJ. Ovarian cancer spheroid shrinkage following continuous exposure to cisplatin is a function of spheroid diameter. Gynecol Oncol. 2017;146(1):161–9.
Sun S, Cai J, Yang Q, Zhao S, Wang Z. The association between copper transporters and the prognosis of cancer patients undergoing chemotherapy: a meta-analysis of literatures and datasets. Oncotarget. 2017;8(9):16036–51.
Saif MW, Heaton A, Lilischkis K, Garner J, Brown DM. Pharmacology and toxicology of the novel investigational agent Cantrixil (TRX-E-002-1). Cancer Chemother Pharmacol. 2017;79(2):303–14.
Kim EM, Jung CH, Kim J, Hwang SG, Park JK, Um HD. The p53/p21 complex regulates Cancer cell invasion and apoptosis by targeting Bcl-2 family proteins. Cancer Res. 2017;77(11):3092–100.
Krempler A, Deckbar D, Jeggo PA, Lobrich M. An imperfect G2M checkpoint contributes to chromosome instability following irradiation of S and G2 phase cells. Cell Cycle. 2007;6(14):1682–6.
Koster R, di Pietro A, Timmer-Bosscha H, Gibcus JH, van den Berg A, Suurmeijer AJ, Bischoff R, Gietema JA, de Jong S. Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J Clin Invest. 2010;120(10):3594–605.
Galanos P, Vougas K, Walter D, Polyzos A, Maya-Mendoza A, Haagensen EJ, Kokkalis A, Roumelioti FM, Gagos S, Tzetis M, et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol. 2016;18(7):777–89.
Theerakitthanakul K, Khrueathong J, Kruatong J, Graidist P, Raungrut P, Kayasut K, Sangkhathat S. Senescence process in primary Wilms' tumor cell culture induced by p53 independent p21 expression. J Cancer. 2016;7(13):1867–76.
Wang MB, Yip HT, Srivatsan ES. Antisense cyclin D1 enhances sensitivity of head and neck cancer cells to cisplatin. Laryngoscope. 2001;111(6):982–8.
Culligan K, Tissier A, Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell. 2004;16(5):1091–104.
Shaikh MF, Morano WF, Lee J, Gleeson E, Babcock BD, Michl J, Sarafraz-Yazdi E, Pincus MR, Bowne WB. Emerging role of MDM2 as target for anti-Cancer therapy: a review. Ann Clin Lab Sci. 2016;46(6):627–34.
Xie N, Ma L, Zhu F, Zhao W, Tian F, Yuan F, Fu J, Huang D, Lv C, Tong T. Regulation of the MDM2-p53 pathway by the nucleolar protein CSIG in response to nucleolar stress. Sci Rep. 2016;6:36171.
Thayer KM, Beyer GA. Energetic landscape of MDM2-p53 interactions by computational mutagenesis of the MDM2-p53 interaction. PLoS One. 2016;11(3):e0147806.
Nihira NT, Ogura K, Shimizu K, North BJ, Zhang J, Gao D, Inuzuka H, Wei W. Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci Signal. 2017;10(466):eaai8026.
Stone S, Jiang P, Dayananth P, Tavtigian SV, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55(14):2988–94.
Cabral VD, Cerski MR, Sa Brito IT, Kliemann LM. p14 expression differences in ovarian benign, borderline and malignant epithelial tumors. J Ovarian Res. 2016;9(1):69.
Wang J, Ding S, Duan Z, Xie Q, Zhang T, Zhang X, Wang Y, Chen X, Zhuang H, Lu F. Role of p14ARF-HDM2-p53 axis in SOX6-mediated tumor suppression. Oncogene. 2016;35(13):1692–702.
Normand G, Hemmati PG, Verdoodt B, von Haefen C, Wendt J, Guner D, May E, Dorken B, Daniel PT. p14ARF induces G2 cell cycle arrest in p53- and p21-deficient cells by down-regulating p34cdc2 kinase activity. J Biol Chem. 2005;280(8):7118–30.
Muer A, Overkamp T, Gillissen B, Richter A, Pretzsch T, Milojkovic A, Dorken B, Daniel PT, Hemmati P. p14(ARF)-induced apoptosis in p53 protein-deficient cells is mediated by BH3-only protein-independent derepression of Bak protein through down-regulation of Mcl-1 and Bcl-xL proteins. J Biol Chem. 2012;287(21):17343–52.
Hemmati PG, Muer A, Gillissen B, Overkamp T, Milojkovic A, Wendt J, Dorken B, Daniel PT. Systematic genetic dissection of p14ARF-mediated mitochondrial cell death signaling reveals a key role for p21CDKN1 and the BH3-only protein puma/bbc3. J Mol Med (Berl). 2010;88(6):609–22.
Peralta S, Clemente P, Sanchez-Martinez A, Calleja M, Hernandez-Sierra R, Matsushima Y, Adan C, Ugalde C, Fernandez-Moreno MA, Kaguni LS, et al. Coiled coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function. J Biol Chem. 2012;287(29):24174–85.
Yin DT, Xu J, Lei M, Li H, Wang Y, Liu Z, Zhou Y, Xing M. Characterization of the novel tumor-suppressor gene CCDC67 in papillary thyroid carcinoma. Oncotarget. 2016;7(5):5830–41.
Wei S, Shang H, Cao Y, Wang Q. The coiled-coil domain containing protein Ccdc136b antagonizes maternal Wnt/beta-catenin activity during zebrafish dorsoventral axial patterning. J Genet Genomics. 2016;43(7):431–8.
Jiang GY, Zhang XP, Zhang Y, Xu HT, Wang L, Li QC, Wang EH. Coiled-coil domain-containing protein 8 inhibits the invasiveness and migration of non-small cell lung cancer cells. Hum Pathol. 2016;56:64–73.
Radulovich N, Leung L, Ibrahimov E, Navab R, Sakashita S, Zhu CQ, Kaufman E, Lockwood WW, Thu KL, Fedyshyn Y, et al. Coiled-coil domain containing 68 (CCDC68) demonstrates a tumor-suppressive role in pancreatic ductal adenocarcinoma. Oncogene. 2015;34(32):4238–47.
Pal D, Wu D, Haruta A, Matsumura F, Wei Q. Role of a novel coiled-coil domain-containing protein CCDC69 in regulating central spindle assembly. Cell Cycle. 2010;9(20):4117–29.
Martin AM, Kanetsky PA, Amirimani B, Colligon TA, Athanasiadis G, Shih HA, Gerrero MR, Calzone K, Rebbeck TR, Weber BL. Germline TP53 mutations in breast cancer families with multiple primary cancers: is TP53 a modifier of BRCA1? J Med Genet. 2003;40(4):e34.
Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14(16):5198–208.
Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Kudo Y, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One. 2010;5(3):e9615.
Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genet. 2009;2:18.
Nozaki M, Tada M, Kashiwazaki H, Hamou MF, Diserens AC, Shinohe Y, Sawamura Y, Iwasaki Y, de Tribolet N, Hegi ME. p73 is not mutated in meningiomas as determined with a functional yeast assay but p73 expression increases with tumor grade. Brain Pathol. 2001;11(3):296–305.
McKenzie PP, Danks MK, Kriwacki RW, Harris LC. P21Waf1/Cip1 dysfunction in neuroblastoma: a novel mechanism of attenuating G0-G1 cell cycle arrest. Cancer Res. 2003;63(13):3840–4.
Khan QA, Vousden KH, Dipple A. Cellular response to DNA damage from a potent carcinogen involves stabilization of p53 without induction of p21(waf1/cip1). Carcinogenesis. 1997;18(12):2313–8.
Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6(9):663–73.
Han J, Goldstein LA, Hou W, Gastman BR, Rabinowich H. Regulation of mitochondrial apoptotic events by p53-mediated disruption of complexes between antiapoptotic Bcl-2 members and Bim. J Biol Chem. 2010;285(29):22473–83.
Yang-Hartwich Y, Soteras MG, Lin ZP, Holmberg J, Sumi N, Craveiro V, Liang M, Romanoff E, Bingham J, Garofalo F, et al. p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene. 2015;34(27):3605–16.
Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395(6698):124–5.
Eymin B, Leduc C, Coll JL, Brambilla E, Gazzeri S. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene. 2003;22(12):1822–35.
Weber HO, Samuel T, Rauch P, Funk JO. Human p14(ARF)-mediated cell cycle arrest strictly depends on intact p53 signaling pathways. Oncogene. 2002;21(20):3207–12.
Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19(6):607–14.
Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–9.
Ichimura K, Bolin MB, Goike HM, Schmidt EE, Moshref A, Collins VP. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60(2):417–24.
Zhao Y, Yao YH, Li L, An WF, Chen HZ, Sun LP, Kang HX, Wang S, Hu XR. Pokemon enhances proliferation, cell cycle progression and anti-apoptosis activity of colorectal cancer independently of p14ARF-MDM2-p53 pathway. Med Oncol. 2014;31(12):288.
Yao D, Wang Y, Xue L, Wang H, Zhang J, Zhang X. Different expression pattern and significance of p14ARF-Mdm2-p53 pathway and Bmi-1 exist between gastric cardia and distal gastric adenocarcinoma. Hum Pathol. 2013;44(5):844–51.
Havrilesky LJ, Alvarez AA, Whitaker RS, Marks JR, Berchuck A. Loss of expression of the p16 tumor suppressor gene is more frequent in advanced ovarian cancers lacking p53 mutations. Gynecol Oncol. 2001;83(3):491–500.
Reles A, Wen WH, Schmider A, Gee C, Runnebaum IB, Kilian U, Jones LA, El-Naggar A, Minguillon C, Schonborn I, et al. Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin Cancer Res. 2001;7(10):2984–97.
Huang X, Yuan X, Chen Z, Liang S. p14ARF enhances cisplatin-induced apoptosis in human osteosarcoma cells in p53-independent pathway. Zhonghua Yi Xue Za Zhi. 2015;95(35):2875–9.
Acknowledgements
The authors would like to thank Dr. Joseph Kwong from The Chinese University of Hong Kong for his technical support.
Funding
National Natural Science Foundation of China (Grant No: 81301827) from State Council in China and Direct Grant for Research (2015.1.040) from The Chinese University of Hong Kong in Hong Kong.
Availability of data and materials
The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
LC and CCW conceived and designed the study. LC collected and analyzed the data. LC drafted the manuscript in collaboration with FZ, CC and CCW. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Primers and PCR conditions used for amplification of p53 (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cui, L., Zhou, F., Chen, C. et al. Overexpression of CCDC69 activates p14ARF/MDM2/p53 pathway and confers cisplatin sensitivity. J Ovarian Res 12, 4 (2019). https://doi.org/10.1186/s13048-019-0479-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13048-019-0479-3