Abstract
Background
4q deletion syndrome is a rare chromosomal disorder that mostly arises de novo. The syndrome is characterized by craniofacial dysmorphism, digital abnormalities, skeletal alterations, heart malformations, developmental delay, growth retardation, Pierre Robin sequence, autistic spectrum and attention deficit-hyperactivity disorder, although not every patient shows the same features. Array comparative genomic hybridization (aCGH) use improves the detection of tiny chromosomal deletions and allows for a better understanding of genotype–phenotype correlations in affected patients. We report the case of a 6-year-old female patient showing mild dysmorphic features, mild mental disabilities and a coagulation disorder as a consequence of a de novo del(4)(q34.1) characterized by aCGH.
Case presentation
A 6-year-old female patient exhibited special craniofacial features, such as backward-rotated ears, upslanted palpebral fissures, broad nasal bridges, anteverted nares, broad nasal alae, smooth philtrums, smooth nasolabial folds, thin lips, horizontal labial commissures, and retrognathia. In the oral cavity, maxillary deformation, a high arched palate, agenesis of both mandibular canines and fusion of two mandibular incisors were observed. She also displayed bilateral implantation of the proximal thumbs, widely spaced nipples, dorsal kyphosis, hyperlordosis, and clitoral hypertrophy. In addition, the patient presented with coagulopathy, psychomotor delay, attention deficit-hyperactivity disorder, and mild mental disability. A chromosomal study showed the karyotype 46,XX,del(4)(q34.1), while an aCGH analysis revealed an 18.9 Mb deletion of a chromosome 4q subtelomeric region spanning 93 known genes.
Conclusion
The clinical manifestations of this patient were similar to those reported in other individuals with 4q deletion syndrome. Although most of the patients with a 4q34 terminal deletion share similarities, variations in phenotype are also common. In general, clinical effects of chromosomal deletion syndromes depend on the length of the deleted chromosomal segment and, consequently, on the number of lost genes; however, in all of these syndromes, there is no simple correlation between the phenotype and the chromosomal region involved, particularly in cases of 4q deletion.
Similar content being viewed by others
Background
Chromosome 4q deletions are classified as interstitial and terminal. Interstitial deletions range from 4q11 to 4q28.3, and terminal deletions span from 4q31.1 [1, 2]. 4q deletion syndrome, due to either interstitial or terminal deletions, is an uncommon chromosomal disorder, with an incidence of nearly 1:100,000 [1], and most cases are de novo [3]. 4q deletions are diagnosed postnatally in equal proportions of males and females; in general, such deletions involve large segments and are detected by GTG banding (G-bands by trypsin and Giemsa) [3]. The principal clinical findings of 4q deletion syndrome are craniofacial dysmorphism (low-set ears, broad nasal bridge, short upturned nose, and micrognathia), digital anomalies, skeletal alterations, heart malformations, developmental delays, growth retardation, Pierre Robin sequence, autistic spectrum disorders and attention deficit-hyperactivity disorder [1, 3]. However, no patient shows all features [1]. Deletions of 4q33 to 4q35 are the least common, and generally, patients present with minor physical anomalies and mild mental disability [4]. Moreover, it has been challenging to determine the karyotype-phenotype correlation in each patient with chromosomal imbalances. However, the use of array comparative genomic hybridization (aCGH) improves the detection of subtle deletions and allows for a better understanding of genotype–phenotype correlations in affected patients [2, 5,6,7,8,9]. Here, we report the case of a 6-year-old female patient with mild dysmorphic features, mild mental disability, coagulopathies and a de novo del(4)(q34.1) deletion encompassing an 18.9 megabase (Mb) loss that includes 93 genes.
Case presentation
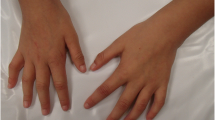
A 6-year-old female patient, the product of the third pregnancy of healthy nonconsanguineous parents (the mother was 20 years old, and the father was 29 at the time the patient was born), presented with poor suckling at birth due to facial muscle hypotonia. At the current physical examination, the patient presented with a height of 114 cm (50th centile), weight of 21.6 kg (50–75th centile), and head circumference of 52 cm (50–75th centile). Craniofacial features included backward-rotated ears, upslanted palpebral fissures, broad nasal bridge, anteverted nares, broad nasal alae, smooth philtrum, smooth nasolabial folds, thin lips, horizontal labial commissures, retrognathia, broad eyebrows, and long eyelashes (Fig. 1a, b). The oral cavity showed maxillary deformation, a high arched palate, agenesis of both canines of the mandible, and fusion between the central and left lateral incisors of the mandible (Fig. 1c, d). The patient had respiratory disorders due to nasal turbinate hypertrophy and hyperplastic tonsils (due to mouth breathing). Moreover, she had widely spaced nipples (Fig. 1e), dorsal kyphosis, hyperlordosis (Fig. 1f), and several nevi (less than 5 mm) in the left supraclavicular fossa, left clavicular region, and right and left hemicollar (Fig. 1e). Her hands showed proximal implantation of the thumbs and fetal fingertip pads (Fig. 1g). Her genitalia showed clitoral hypertrophy (20 mm) (Fig. 1h). Furthermore, the patient had coagulopathies presenting as recurrent hematomas and a long clotting time, as well as excessive bleeding during tooth loss. Her prothrombin time (PT) and activated partial thromboplastin time (aPTT) were 13 s and 44.5 s, respectively (reference values: 12–14 s and 35–43 s, respectively). Other anomalies included psychomotor delay, attention deficit-hyperactivity disorder, anxiety, sphincter control absence and mild mental disability.
Clinical findings of the patient. a Upslanted palpebral fissures, broad nasal bridge, anteverted nares, broad nasal ala, smooth philtrum, smooth nasolabial folds, thin lips, horizontal labial commissures, and broad eyebrows. b Ears rotated backwards, retrognathia, and long eyelashes. c High arched palate. d Canines agenesis and fusion between the central and left lateral incisor of the mandible. e Nevi and widely spaced nipples. f Dorsal kyphosis and hyperlordosis. g Proximal implantation of the thumbs. h Clitoral hypertrophy
Results
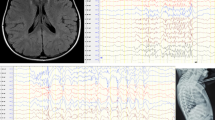
A chromosomal analysis of cultured peripheral blood lymphocytes from the patient was performed by the GTG-banding method at a resolution of 500–550 bands [10] and revealed a karyotype 46,XX,del(4)(q34.1) (Fig. 2a). Both parents were chromosomally normal. A FISH study with a mixture of commercial subtelomeric 4p/4q probes (Cytocell: LPT 04PG and LPT 04QR) confirmed the absence of one copy of subtelomeric 4q sequences on one chromosome 4 (Fig. 2b). To determine the genomic imbalance, aCGH on the proband was performed using CytoScan™ Technology (Thermo Fisher Scientific Inc). Reactions of digestion, ligation, PCR, purification of PCR products, quantification, fragmentation, labeling, matrix hybridization, washing, staining, and scanning arrays were performed according to the manufacturer’s instructions. Data were analyzed with ChAS 4.0 software. Interpretation of the results was performed using the following databases: Ensembl Resources, Database of Genomic Variants, Cytogenomics Array Group Copy Number Variant, Online Mendelian Inheritance in Man (OMIM), University of California Santa Cruz Database, ClinGen, and ClinVar databases. The analysis revealed the following 18.9 Mb deletion of a chromosome 4q region containing approximately 93 genes: arr[GRCh38] 4q34.1q35.2(171,135,044_190,036,318)×1 dn (Fig. 2c). Hence, the integrated proposita’s karyotype was 46,XX,del(4)(q34.1)[20].ish 4pter(subtel(4p)×2),4qter(subtel(4q)×1)[15].arr[GRCh38]4q34.1q35.2(171,135,044_190,036,318)×1 dn.
Chromosomal and aCGH studies in the patient. a Selected GTG-banded chromosomes. Normal chromosome 4 is on the left and chromosome 4 with the deletion is on the right. b FISH study with a mixture of subtelomeric 4p/4q molecular probes, labeled in green and red, respectively. The subtelomeric 4q red signal is absent from the deleted chromosome 4. c aCGH image exhibit an 18.9 Mb terminal deletion of the chromosome segment 4q34.1q35.2 (171,135,044-190,036,318, GRCh38/hg38). The deleted chromosomal region is delimited by the red rectangle
Discussion and Conclusions
The patient’s net genomic imbalance was an 18.9 Mb deletion spanning approximately 93 known genes, from MIR6082 to FRG2, including 44 genes recorded in the OMIM database. Among them, HPGD, VEGFC, AGA, TENM3, TRAPPC11, CASP3, PRIMPOL, SLC25A4, UFSP2, TLR3, CYP4V2, DFNA24, PDLIM3, SORBS2, KLKB1, F11, and FRG1 are mutated in some disorders [11]. Other genes in this region, such as HAND2, PDLIM3, and SORBS2, have been implicated in congenital heart defects [2], particularly HAND2 (chr4:173,526,091-173,530,229, GRCh38), of which mutations have been transmitted in a pattern of dominant inheritance [12].
Our patient presented with clinical features typical of 4q terminal deletions, as well as clitoral hypertrophy and idiopathic coagulopathy, but not cardiopathy. Vona et al. [2] reported the case of a young male patient with a 4q35.1q35.2 deletion (chr4:184,046,156-190,901,117, GRCh37; chr4:183,125,003-189,979,962, GRCh38) affecting 6.9 Mb, who, in addition to heart defects, hearing impairment, cleft palate, and bilateral cryptorchidism, at age five developed coagulation factor XI deficiency. Moreover, Guéguen et al. [13] reported the cases of a woman and two of her children with a 4q34.2 deletion spanning ~ 7 Mb (chr4:182,720,115-191,044,276, GRCh37; chr4:182,798,962-190,214,555, GRCh38) with a variable bleeding phenotype. These are the only known patients with 4q terminal deletion and a coagulation disorder, which could be related to the loss of two genes of the coagulation pathway, KLKB1 (chr4:186,227,507-186,258,471, GRCh38) and F11 (chr4:186,265,945-186,288,780, GRCh38), in particular F11, of which some mutations have shown dominant inheritance in patients with bleeding tendency [14]. To the best of our knowledge, clitoral hypertrophy has not been previously associated with 4q terminal deletions.
Several cases of terminal deletions overlapping 4q34 or 4q35 have been reported. Although many of the affected patients share similar clinical presentations, variations in phenotype are common. While our patient did not present with heart disease, she shares multiple features with these other patients, in particular facial dysmorphism, nasal anomalies, a high arched palate, digital anomalies, psychomotor delay, and mental disability (all these clinical features are summarized in Table 1) [4, 5, 7, 15,16,17,18,19]. In this regard, Rossi et al. [5] described a young woman with a sporadic 4q34.1q35.2 deletion encompassing 16.44 Mb (chr4:174,685,919-191,121,195, hg18; chr4:173,528,193-189,963,046, GRCh38, from genes HAND2 to FRG1) who was diagnosed with learning disabilities, Pierre Robin sequence, and heart defects, such as atrial septal and Ebstein anomalies. A further de novo 4q34.1q35.2 deletion spanning 17.4 Mb (chr4:172,977,872-190,351,861, hg18; chr4:171,820,146-189,193,713, GRCh38) was detected in a young boy who presented with tetralogy of Fallot, right aortic arch, and facial dysmorphism, resembling 22q11.2 deletion syndrome [6]. Similarly, Tsai et al. [4] described a child with a de novo 4q34.2 terminal deletion who presented with cardiac defects, a cleft palate, learning difficulties, and right fifth finger anomalies, consistent with velocardiofacial syndrome, but negative for the 22q11.2 deletion. Connel et al. [20] reported a girl with 4q34 deletion who, in addition to facial dysmorphism, digital anomalies, and heart defects, also presented with bilateral optic disk swelling. Bendavid et al. [21] identified a woman and her daughter carrying a 4q34.3 terminal deletion distal to the AGA gene [at least from TENM3 (chr4:182,243,402-182,803,024, GRCh38; approximately 8.7 Mb)] who exhibited different phenotypes. While the mother showed a cardiac defect, the daughter had congenital absence of the upper vagina and uterus. Marci et al. [22] described a female patient and her child carrying the same 4q34.3 deletion but with different phenotypes. While the mother had nonobstructive cor triatriatum sinister, the son presented with tetralogy of Fallot. Descartes et al. [18] reported a maternal 4q34.2 terminal deletion in two siblings with growth retardation, intellectual disability, and craniofacial alterations, but their mother only showed craniofacial alterations. Moreover, by aCGH studies, Youngs et al. [7] detected a 4q35.2 interstitial microdeletion of nearly 1.2 Mb (chr4:187.47-188.66, GRCh37; chr4:186,548,846-187,738,846, GRCh38), involving MTNR1A, FAT1, and F11, in an autistic boy who also showed congenital cardiopathy, psychomotor delay, facial dysmorphism, and mental disability. Similarly, through microarrays, Shao et al. [23] identified six patients with 4q35 deletion who had multiple congenital anomalies, with either dysmorphism, developmental delay/mental disability, or seizure disorders. They also reported two patients with 4q35 deletion and minor alterations. One had only dysmorphic features, while the other showed respiratory distress syndrome and asthma exacerbation. Strehle et al. [9] studied a patient with almost all clinical characteristics of 4q deletion syndrome and found a deletion of nearly 465 kb in 4q35.1 (chr4: 186,770,069-187,234,800, hg18; chr4:185,611,921-186,076,652, GRCh38, case 20); they proposed that this region is critical for the expression of this condition. Nevertheless, 4q35 terminal deletions have also been found in patients who only have autism or psychiatric diseases. Chien et al. [8] described a boy with autism and 4q35.1q35.2 deletion affecting approximately 6.8 Mb (chr4:183,904,000-190,720,000, hg18; chr4:182,745,853-189,561,852, GRCh38), and Pickard et al. [24] identified a man with a schizoaffective disorder and mental disability who had a 4q35.2 deletion, spanning approximately 3.0 Mb (nearly chr4:187,214,555-190,214,555, GRCh38). Additionally, several individuals with 4q34 or 4q35 deletions and a normal phenotype have been reported [13, 25–29, Table 2].
Interestingly, it should be noted that although Pierre Robin sequence is commonly associated with 4q deletion, only one patient in our review showed this feature, which could indicate that 4q34 → qter is not a critical region for Pierre Robin sequence. Since not every individual with deletions involving the HAND2 gene presents with congenital heart defects (neither our patient nor the patient described by Huang et al. [28] presented with such defects) and because some patients with distal deletions of HAND2 do show those defects [2, 4, 5, 7, 21, 22, 28], it is probable that other genes in this region play important roles in heart development, as is the case for the SORBS2 gene [30], located at 4q35.1. Last, although clinical effects generally depend on the length of the chromosomal segment and, consequently, on the number of genes deleted, there is no direct correlation between the phenotype and the chromosomal region involved. Even though the reasons why similar deletions result in great phenotypical heterogeneity or even in normal phenotypes are not currently known, environmental variables interacting with the genetic background, some modifier genetic variants and/or epigenetic changes could play key roles that contribute to such heterogeneity [31]. It is obvious that further research is necessary to clarify this issue.
Availability of data and materials
Data generated or analyzed during this study are included in this published article.
Abbreviations
- aCGH:
-
Array comparative genomic hybridization
- Mb:
-
Megabase
- GTG-banding:
-
G-bands by trypsin using Giemsa
- PT:
-
Prothrombin time
- aPTT:
-
Activated partial thromboplastin time
References
Strehle EM, Gruszfeld D, Schenk D, Mehta SG, Simonic I, Huang T. The spectrum of 4q- syndrome illustrated by a case series. Gene. 2012;506:387–91.
Vona B, Nanda I, Neuner C, Schroder J, Kalscheuer VM, Shehata-Dieler W, et al. Terminal chromosome 4q deletion syndrome in an infant with hearing impairment and moderate syndromic features: review of literature. BMC Med Genet. 2014;15:72.
Strehle EM, Bantock HM. The phenotype of patients with 4q syndrome. Genet Couns. 2003;14:195–205.
Tsai CH, Van Dyke DL, Feldman GL. Child with velocardiofacial syndrome and del (4)(q34.2): another critical region associated with a velocardiofacial syndrome-like phenotype. Am J Med Genet. 1999;82:336–9.
Rossi MR, DiMaio MS, Xiang B, Lu K, Kaymakcalan H, Seashore M, et al. Clinical and genomic characterization of distal duplications and deletions of chromosome 4q: study of two cases and review of the literature. Am J Med Genet A. 2009;149A:2788–94.
Cuturilo G, Menten B, Krstic A, Drakulic D, Jovanovic I, Parezanovic V, et al. 4q34.1-q35.2 deletion in a boy with phenotype resembling 22q112 deletion syndrome. Eur J Pediatr. 2011;170:1465–70.
Youngs EL, Henkhaus RS, Hellings JA, Butler MG. 12-year-old boy with a 4q35.2 microdeletion and involvement of MTNR1A, FAT1, and F11 genes. Clin Dysmorphol. 2012;21:93–6.
Chien WH, Gau SS, Wu YY, Huang YS, Fang JS, Chen YJ, et al. Identification and molecular characterization of two novel chromosomal deletions associated with autism. Clin Genet. 2010;78:449–56.
Strehle EM, Yu L, Rosenfeld JA, Donkervoort S, Zhou Y, Chen TJ, et al. Genotype-phenotype analysis of 4q deletion syndrome: proposal of a critical region. Am J Med Genet A. 2012;158:2139–51.
ISCN 2020: An International system for human cytogenomic nomenclature. McGowan-Jordan J, Hastings Simons RJ, Moore S (eds). Cytogenet Genome Res. 2020;160:341–503.
Online Mendelian Inheritance in Man, OMIM. An Online Catalog of Human Genes and Genetic Disorders. 2021. Available from: https://omim.org/. Accessed 28 May 2021.
Cohen ASA, Simotas C, Webb BD, Shi H, Khan WA, Edelmann L, et al. Haploinsufficiency of the basic helix-loop-helix transcription factor HAND2 causes congenital heart defects. Am J Med Genet. 2020;182A:1263–7.
Guéguen P, Chauvin A, Quémener-Redon S, Pan-Petesch B, Férec C, Abgrall JF, et al. Revisiting the molecular epidemiology of factor XI deficiency: nine new mutations and an original large 4qter deletion in western Brittany (France). Thromb Haemost. 2012;107:44–50.
Online Mendelian Inheritance in Man, OMIM. An Online Catalog of Human Genes and Genetic Disorders. 2021. Available from: https://omim.org/entry/264900. Accessed 28 May 2021.
Lin AE, Garver KL, Diggans G, Clemens M, Wenger SL, Steele MW, et al. Interstitial and terminal deletions of the long arm of chromosome 4: further delineation of phenotypes. Am J Med Genet. 1988;31:533–48.
Vogt J, Ryan E, Tischkowitz MD, Reardon W, Brueton LA. The tale of a nail sign in chromosome 4q34 deletion syndrome. Clin Dysmorphol. 2006;15:127–32.
González MS, González García N, Tasé Vila D, SuárezMayedo U, Calixto Robert Y. Deletion of the long arm of the 4(q34-qter) chromosome. Report of cytogenetic finding in one case. Rev Cub Gen Comunit. 2014;8:55–8.
Descartes M, Keppler-Noreuil K, Knops J, Longshore JW, Finley WH, Carroll AJ. Terminal deletion of the long arm of chromosome 4 in a mother and two sons. Clin Genet. 1996;50:538–40.
Caliebe A, Waltz S, Jenderny J. Mild phenotypic manifestations of terminal deletion of the long arm of chromosome 4: clinical description of a new patient. Clin Genet. 1997;52:116–9.
Connel P, Brosnahan D, Dunlop A, Reardon W. Bilateral optic disk swelling in the 4q34 deletion syndrome. J AAPOS. 2007;11:516–8.
Bendavid C, Pasquier L, Watrin T, Morcel K, Lucas J, Gicquel I, et al. Phenotypic variability of a 4q34-qter inherited deletion: MRKH syndrome in the daughter, cardiac defect and Fallopian tube cancer in the mother. Eur J Med Genet. 2007;50:66–72.
Marci M, Guarina A, Castiglione MC, Sanfilippo N. Different cardiac anomalies in mother and son with 4q-syndrome. Case Rep Genet. 2015;2015:932651
Shao L, Shaw CA, Lu XY, Sahoo T, Bacino CA, Lalani SR, et al. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: a study of 5,380 cases. Am J Med Genet A. 2008;146A:2242–51.
Pickard BS, Hollox EJ, Malloy MP, Porteous DJ, Blackwood DH, Armour JA, et al. A 4q35.2 subtelomeric deletion identified in a screen of patients with co-morbid psychiatric illness and mental retardation. BMC Med Genet. 2004;13:21.
Bateman MS, Mehta SG, Willatt L, Selkirk E, Bedwell C, Zwolinski S, et al. A de novo 4q34 interstitial deletion of at least 93 Mb with no discernible phenotypic effect. Am J Med Genet A. 2010;152A:1764–9.
Yakut S, Clarck OA, Sanhal C, Nur BG, Mendilcioglu I, Karauzum SB, et al. A familial interstitial 4q35 deletion with no discernible clinical effects. Am J Med Genet A. 2014;167A:1836–41.
Balikova I, Menten B, de Ravel T, Le Caignec C, Thienpont B, Urbina M, et al. Subtelomeric imbalances in phenotypically normal individuals. Hum Mutat. 2007;28:958–67.
Huang T, Lin AE, Cox GF, Golden WL, Feldman GL, Ute M, et al. Cardiac phenotypes in chromosome 4q-syndrome with and without a deletion of the Dhand gene. Genet Med. 2002;4:464–7.
Ravnan JB, Tepperberg JH, Papenhausen P, Lamb AN, Hedrick J, Eash D, et al. Subtelomere FISH analysis of 11,688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet. 2006;43:478–89.
Liang F, Wang B, Geng J, You G, Fa J, Zhang M, et al. SORBS2 is a genetic factor contributing to cardiac malformation of 4q deletion syndrome patients. Elife. 2021;10: e67481. https://doi.org/10.7554/eLife.67481.
Meza-Espinoza JP, Sáinz González E, León-León CJN, Arámbula-Meraz E, Contreras-Gutiérrez JA, García-Magallanes N, et al. Report of trisomy 2q34-qter and monosomy 4q35.2-qter in a child with mild dysmorphic syndrome and karyotype 46,XY,der(4)t(2;4)(q34;q35.2)pat. Mol Cytogenet. 2020;13:17.
Acknowledgements
We thank the patient’s parents for their collaboration and to María de Lourdes Carbajal for her review of the writing. Dr Christian NJ León-León helped in clinical examination and dental medicine student Julissa Karina Ibarra Miranda supported in the oral clinical examination.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
JPME: Data collection and article drafting. JACG: Clinical review of the urinary tract and article writing. EAM, NGM, MPP: Collection of samples, making, and interpretation microarrays studies. JRGG, MGDQ: Performance and interpretation of FISH studies and intellectual deliver during the drafting. JMM: Clinical examination. JBP: Patient recruitment and oral inspection, referral to the genetics department. VJPC: Conception and design of the study, drafting, acquisition and interpretation of data. All the authors contributed to the critical review and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by our ethical committee and the authors have no ethical conflicts to disclose.
Consent for publication
Patient’s parents authorized the use of biological and photographic material for research purposes through informed consent.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Meza-Espinoza, J.P., Contreras-Gutiérrez, J.A., Arámbula-Meraz, E. et al. Cytogenomic characterization of a de novo 4q34.1 deletion in a girl with mild dysmorphic features and a coagulation disorder. Mol Cytogenet 14, 43 (2021). https://doi.org/10.1186/s13039-021-00564-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-021-00564-z