Abstract
Background
Non-invasive prenatal testing (NIPT) as alternative screening method had been proven to have very high sensitivity and specificity for detecting common aneuploidies such as T21, T18, and T13, with low false positive and false negative rates. Unfortunately, recent studies suggested that the NIPT achieved lower accuracy in sex chromosomal aneuploidies (SCAs) detection than autosomal aneuploidies detection. BGISEQ-500 powered by Combinatorial Probe-Anchor Synthesis (CPAS) and DNA Nanoballs (DNBs) technology that combined linear amplification and rolling circle replication to reduce the error rate while enhancing the signal. Therefore, NIPT based on CPAS might be a good method for SCAs screening in routine clinical practice. In the study, we intended to evaluate the clinical utility of NIPT based on CPAS on screening for fetal SCAs.
Results
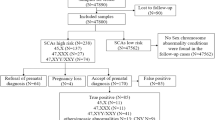
A total of 570 pregnant women were included in the retrospective study. Maternal blood samples were collected for NIPT; amniocentesis was performed on all pregnant women. NIPT was carried out by BGISEQ-500 sequencing platform based on CPAS. Karyotype analysis of amniotic cells was performed by standard G-banding techniques. 43 out of the total 570 pregnant women tested by NIPT showed fetal SCAs (19 of 45,X, 12 of 47,XXY, 10 of 47,XXX, and 2 of 47,XYY). The following amniocentesis confirmed that 26 cases were true positive (7 of true positive 45,X, 9 of true positive 47,XXY, 9 of true positive 47,XXX as well as 1 of 47,XYY) and the positive predictive value (PPV) for fetal SCAs was 60.47%. In addition, the PPV of advanced maternal age group (67.74%) was higher than the other indications group (45.45%) or serological screening high-risk /critical-risk group (0%).
Conclusions
NIPT based on CPAS could be a potential method for SCAs screening. However, it still had high false positive rates, especially for 45,X. The pregnant women with fetal SCAs detected by NIPT, especially those with non-age-related prenatal diagnostic indications, should be advised to accept invasive prenatal karyotype analysis.
Similar content being viewed by others
Background
Since cell-free fetal DNA (cffDNA) was detected in cell-free DNA (cfDNA) of pregnant women plasma by Lo et al. in 1997 [1], comparison of chromosome dosage distribution of cffDNA between patient and control plays an increasingly important role in fetal aneuploidy diagnosis [2]. NIPT calculated the risk of fetal chromosomal aneuploidies by detecting cffDNA circulating in maternal plasma with the next-generation sequencing technology [3]. The NIPT as alternative screening method had been proven to have > 99% sensitivity and a false-positive rate of < 0.1% for autosomal aneuploidies such as trisomy 21 (T21, Down syndrome), trisomy 18 (T18), trisomy 13 (T13) [4]. Unfortunately, recent studies suggested that the NIPT achieved lower accuracy in SCAs detection than autosomal aneuploidies detection [5, 6]. One of the most important reasons is that SCAs detection is highly dependent on fetus sex prediction [2]. SCAs such as Turner (45,X), Klinefelter (47,XXY), Triple X syndrome (47,XXX) and Jacob syndromes (47,XYY) affected approximately one in 1000 births [7], early interventions such as hormone therapy and hormone replacement therapy had been shown to improve the outcomes for babies with 45,X syndrome or 47,XXY syndrome [8]. So prenatal screening and prenatal diagnosis about SCAs were critical for outcome preparation and timely treatment [9].
NIPT had been rapidly used in clinical in China since 2011 [10]. The Women’s Hospital School of Medicine, Zhejiang University had validated and implemented NIPT successfully on October 2012, and obtained the first batch of National Clinical Trial Unit qualification in 2015. BGISEQ-500 is a recently launched high-throughput sequencing solution, powered by Combinatorial Probe-Anchor Synthesis (CPAS) and improved DNA Nanoballs (DNBs) technology. The CPAS chemistry works by incorporating a fluorescent probe to a DNA anchor on the DNB, followed by high-resolution digital imaging. This combination of linear amplification and DNB technology reduces the error rate while enhancing the signal [11,12,13]. Therefore, NIPT based on CPAS might be a good method for SCAs screening in routine clinical practice. In the study, we intended to evaluate the clinical utility of NIPT based on CPAS on screening for fetal SCAs.
Materials and methods
Subjects
A total of 570 pregnant women who visited the Women’s Hospital School of Medicine, Zhejiang University for prenatal counseling from May 2017 to October 2017 were included in the retrospective study. The study was carried out under the approvement by the Hospital Ethics Committee of the Women’s Hospital School of Medicine, Zhejiang University. All 570 pregnant women had received amniocentesis and karyotype analyzing at Women’s Hospital School of Medicine, Zhejiang University.
The indications for NIPT were as follows:
Maternal serological screening high-risk value (T21 ≥ 1/270, T18 ≥ 1/350) or critical-risk value (1/1000 ≤ T21 < 1/270, 1/1000 ≤ T18 < 1/350), advanced maternal age (35 years or older at the expected date of confinement), abnormal fetal structure identified by ultrasound, adverse pregnancy history or pregnant women involving “anxiety” who voluntarily required NIPT.
The exclusion criteria were in accordance with the rules of Chinese government and were as follows: twin or multiple pregnant women, either parent of the fetus with chromosomal abnormality, pregnant women who undergo embryo transplantation or stem cell therapy, and pregnant women who had a history of immunotherapy within 1 month or allogeneic blood transfusion within 1 year.
Among the 570 pregnant women analyzed by NIPT, 49 were found to have autosomal aneuploidies, including 3 of T13 (3/570), 8 of T18 (8/570) and 38 of T21 (38/570). 43 were found to have SCAs (43/570), including 19 of 45,X (19/570), 12 of 47,XXY (12/570), 10 of 47,XXX (10/570), and 2 of 47,XYY (2/570).
Collection and treatment of blood samples
Maternal peripheral blood samples (5 ml) were collected in EDTA tubes, fully mixed, stored temporarily in 4 °C refrigerator. Samples were excluded if hemolysis or storage beyond 8 hours before plasma separation. The blood sample was treated as follows: centrifuged at 4 °C, 1600 g for 10 min and the plasma was collected carefully and dispensed into 2.0 ml Eppendorf tubes. The plasma was centrifuged again at 4 °C, 16,000 g for another 10 min. The upper plasma was carefully divided into new 2.0 ml Eppendorf tubes and each contained approximately 600 ml plasma, − 80 °C refrigerator to save. The plasma should avoid repeated freezing and thawing before experiment.
Non-invasive prenatal testing (NIPT)
The DNA extraction, library construction, and sequencing were performed according to the standard protocol of the Women’s Hospital School of Medicine, Zhejiang University of Human Molecular Genetics Guidelines. 200 μl maternal plasma was used for cffDNA extraction by BGISP-300 (BGI, Shenzhen, China) with Nucleic Acid Extraction Kit (BGI, Shenzhen, China). The next step after DNA extraction was end-repair by adding end-repair enzymes with the following cycle conditions: 37 °C for 10 min and 65 °C for 15 min, then followed by adaptor ligation at 23 °C for 20 min with label-adaptor and ligase. After end-repair and adaptor ligation, PCR was used to amplify DNA to desired concentration with the following cycle conditions: 98 °C for 2 min, then 12 cycles at 98 °C for 15 s, 56 °C for 15 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. The DNA amplification products were quantified by Qubit® 2.0 (Life Tech, Invitrogen, USA) using QubitTM dsDNA HS Assay Kits (Life Tech, Invitrogen, USA) and the concentration ≥ 2 ng/μl was regarded as qualified standards. The volume was calculated according to the concentration of each sample, and each sample of the same mass was mixed by pooling. The DNA double strands were thermally denatured into single strand after pooling, and then the cyclic buffer and the ligase were added to make DNA circles by the cyclization reaction. DNA circles were used to make DNBs by rolling circle replication (RCR). The concentration of DNBs was quantified by Qubit® 2.0 using QubitTM ssDNA Assay Kits (Life Tech, Invitrogen, USA) and the DNBs concentration in the range of 8 ng/μl-40 ng/μl was considered as appropriate concentration. The DNBs were loaded onto chips and sequenced on BGISEQ-500 sequencing platform (BGI, Shenzhen, China). Fetal chromosome aneuploidies (T21, T18, and T13) detection kit (Combinatorial Probe-Anchor Synthesis Sequencing Method) (BGI, Shenzhen, China) was used for library construction and Sequencing was performed using Universal Reaction Kit for Sequencing (Combinatorial Probe-Anchor Synthesis Sequencing Method) (BGI, Shenzhen, China). Any sample that failed to meet the quality control criteria was reported as detection failure by NIPT. Z-score set the range of − 3 and 3 as the threshold to evaluate the risk of chromosomal aneuploidies. If Z-score > 3 or Z-score < − 3, the sample was classified as high-risk of chromosomal aneuploidies. If Z-score was between − 3 to 3, the sample was classified as low risk.
Amniocentesis and fetal karyotyping
Amniocentesis was performed at gestational age of 19+ 0 to 22+ 6 weeks. All pregnant women signed informed consents before the invasive procedure. Amniocentesis was performed by the certificated physicians under ultrasound guidance. The initial 2 ml of amniotic fluid was discarded and the other 20 ml of amniotic fluid was collected for karyotype analysis. Amniotic fluid cells were cultured according to the standard protocol of the Women’s Hospital School of Medicine, Zhejiang University of Human Cytogenetics Guidelines. Chromosome analysis with targeted 400-band level were performed and at least 30 metaphases were counted and 5 metaphases were analyzed for each sample by using GSL-120 high-throughput automatic chromosome scanning platform (Leica, German) after G-banding. If there were different cell lineages in the same patient, the count was increased to 50–100 metaphases to establish the mosaicism. Chromosomal abnormalities were described according to the criteria established by the International System for Human Cytogenetic Nomenclature (ISCN, 2016).
Statistics
The statistics were calculated treating the invasive procedure results as the gold standard, which included the false positive rate (FPR), false negative rate (FNR), sensitivity (SEV), specificity (SPC) as well as positive predict value (PPV) and negative predict value (NPV) in the study. The 95% confidence intervals (CI) were calculated on the bases of a standard normal distribution by Clopper-Pearson method.
Results
In the present study, a total of 570 pregnant women were enrolled for the NIPT trial according to different prenatal diagnosis indications, including 328 with advanced maternal age, 78 with serological screening high-risk, 2 with serological screening critical-risk, and 162 with other indications (ultrasound fetal structural abnormalities or ultrasound soft indexes abnormality, adverse pregnancy history and pregnant women who voluntarily required NIPT). The proportion of the different prenatal diagnosis indications was listed in Table 1.
After NIPT detection, 43 pregnant women had abnormal SCAs results as follows: 45,X in 19/570; 47,XXY in 12/570; 47,XXX in 10/570 and 47,XYY in 2/570. The following amniocentesis results confirmed that there were 26 cases of true SCAs (26/570), including 7 true positive of 45,X (7/570), 9 true positive of 47,XXY (9/570), as well as 9 true positive of 47,XXX (9/570) and one of 47,XYY (1/570). All 49 pregnant women (T13 in 3/570, T18 in 8/570 and T21 in 38/570) with autosomal aneuploidies detected by NIPT had the same results with the amniocentesis. The SCAs results of NIPT and amniocentesis were listed in Table 2. Further analysis showed that the overall FPR of SCAs in the study was 3.13%, the FPR of 45,X, 47,XXY, 47,XXX, and 47,XYY was 2.13, 0.53, 0.18, and 0.18%, respectively. The number of false negative was 0, and the FNR of SCAs in the study was 0% (Table 3).
The SEV, SPC, NPV, and PPV of SCAs by NIPT based on BGISEQ-500 were listed in Table 4. The overall SEV was 100% (95% CI: 86.77–100.00%), SPC was 96.88% (95% CI: 95.04–98.17%) and NPV was 100%. The overall PPV was 60.47% (95% CI: 48.93–70.95%), and the PPV for 45,X, 47,XXY, 47,XXX, and 47,XYY was 36.84% (95% CI: 25.00–50.52%), 75.00% (95% CI: 49.25–90.27%), 90.00% (95% CI: 55.95–98.46%%), and 50.00% (95% CI: 12.37–87.63%), respectively.
We further classified the 43 pregnant women with fetal SCAs according to different prenatal diagnostic indications (Table 5). 31 patients were from advanced maternal age group, one patient was from serological screening high-risk / critical-risk group and 11 patients from other indications group. The true positive rate of SCAs in advanced maternal age group was 67.74%, and the positive rate of 45,X, 47,XXY, 47,XYY, and 47,XXX in advanced maternal age group was 45.45, 72.73, 50, and 100%, respectively. And the overall positive rate in other group was 45.45%, and the positive rate of 45,X, 47,XXY, 47,XXX in other indications group was 25, 100, 100%, respectively. The positive rate of SCAs in serological screening high-risk / critical-risk value group was 0%.
Discussion
Massive Parallel Genome Sequencing (MPGS) (also called as next-generation sequencing, NGS) had been widely used in NIPT for years [5, 14]. NIPT as a successful application in routine clinical practice has been widely used to detect trisomy 21, 18, 13 and SCAs [15, 16]. The result of fetal sex prediction was dependent on SCAs detection. However, the current data processing method has some limitations to achieve higher accuracy. In the present study, we investigated the clinical utility of NIPT based on CPAS that applied DNBs technology for sequencing library construction and CPAS for massively parallel DNA sequencing, and found that NIPT based on BGISEQ-500 sequencing platform was suitable for SCAs detection.
There were 43 pregnant women with fetal SCAs shown by NIPT, and the overall PPV was 60.47% (95%CI 48.93–70.95%), which was higher than some existing researches [17,18,19]. The higher PPV reflected a lower omission diagnose rate and a higher detection rate of target disease based on BGISEQ-500 sequencing platform. The source of the patient might be an important factor. As the largest obstetrics and gynecology hospital in Zhejiang Province, the majority of patients were relatively higher risk individuals. In addition, the small number of samples tested might also be an important contributing factor. For example, there were only two cases of 47,XYY, which would result in wide 95% CI (12.37–87.63%) and low statistical power. The PPV range from 47,XXX (90, 95%CI 55.95–98.46%), 47,XXY (75.00, 95%CI 49.25–90.27%), 47,XYY (50, 95%CI 12.37–87.63%) to 45,X (36.84, 95%CI 25.00–50.52%). The PPV for detecting 45,X was the lowest, indicating the NIPT based on BGISEQ-500 sequencing platform for 45,X still had high false positive rates and the accuracy needed to be further improved. However, the PPV for detecting 45,X was 36.84%, which was relative higher compared with other studies [19]. Therefore, NIPT that based on BGISEQ-500 sequencing platform for screening fetal SCAs still need more improvement. Based on our study, the prevalence of true SCAs in population was 4.56% (26/570), which would avoid 95.46% of unnecessary invasive procedures. However, compared with amniocentesis, NIPT based on the BGISEQ-500 sequencing platform for SCAs still had high false positive rates, especially for Turner’s syndrome. Therefore, the pregnant women with fetal SCAs by NIPT should be advised to accept invasive prenatal karyotype analysis.
At present, the NIPT platform was highly sensitive and specific for screening autosomal trisomy [20, 21]. However, the application of prenatal diagnosis for SCAs was still in constant exploration [21, 22]. According to our results, the false positive rate and the false negative rate for SCAs was 3.13 and 0%, respectively. In addition to the limitations of noninvasive screening techniques, the source of cffDNA is another important factor to lead to inconsistent results between NIPT and traditional invasive diagnosis. It was well known that the presence of cffDNA circulating in maternal plasma was derived from the placental trophoblast [23]. These discordant cases are predominantly due to confined placental mosaicism (CPM). CPM can lead to false positive (abnormal cells in placenta but normal in fetus); or false negative results (abnormal cells in the fetus but not placenta) [24]. In addition, true fetal mosaicism can also lead to discordant NIPT results, which has been predicted to occur in 1/107 cases [24]. Maternal etiology for discordant results can arise if the mother is a mosaic herself, this is particularly the case when using NIPT to screen for SCAs [25].
In the study, the sensitivity and negative predict value of all kinds of SCAs were 100%, suggesting that NIPT can identify more positive cases and can effectively reduce the chance of misdiagnosis in population. The comparisons of specificity are as follows: 47,XYY (99.82, 95%CI 99.02–100.00%) = 47,XXX (99.82, 95%CI 99.01–100.00%) > 47,XXY (99.47, 95%CI 98.45–99.89%) > 45,X (97.87, 95%CI 96.31–98.89%). The overall sensitivity and specificity were 100% (95%CI 86.77–100.00%) and 96.88% (95% CI 95.04–98.17%), which was relative higher compared with some existing researches [26, 27]. This indicated that the BGISEQ-500 sequencing platform had certain advantages in detecting SCAs, but whether the BGISEQ-500 sequencing platform was superior to other sequencing platforms still needed more discussion. The comparisons of accuarcy are as follows: 47,XYY (99.82, 95%CI 99.03–100.00%) > 47,XXY (99.47, 95% CI 98.47–99.89%) > 47,XXX (97.89, 95%CI 96.35–98.91%) = 45,X (97.89, 95%CI 96.35–98.91%), the high accuracy provided wide application prospect in the field of scientific research and clinical testing.
This finding makes us think further about the relationship between the specificity and the PPV of NIPT analyses. The PPV of NIPT is the highest for T21 and is much lower for other aneuploidies [28,29,30,31,32]. PPV depends not only on the sensitivity and specificity of the assay, but also on the prevalence of the disease [33]. In addition, our study was designed to compare the results of NIPT screening and karyotype analysis for fetal SCAs only in patients who had results from both tests and not to compare the performance in actual clinical practice. We excluded the “no call” results including some cases who did not complete NIPT screening or patients without karyotype results. The prevalence was based on evaluating 570 pregnant women in this manuscript, the small numbers in this cohort prevented an accurate assessment of the sensitivity and PPV of the methods.
Due to the relatively low amount of cffDNA, fetal fraction of Cell-Free DNA is a prerequisite for successful NIPT testing. Low fetal fraction was another cause of discordant inconsistent results between NIPT and traditional invasive diagnosis, and higher concentration of fetal DNA associated with more accurate aneuploidy results [34]. The method of sample collection is important for maintaining fetal fraction. More cffDNA is found in the plasma of maternal blood instead of the cellular fraction [35]. It is important to maintain maternal cells integrity when collecting samples for cfDNA extraction. Inappropriate sample collection leads to reduced the detection of fetal fraction. If using of K3EDTA tube sample should be processed within 8 h, otherwise cell-stabilizing tubes should be used [36]. A total of 570 plasma samples were analyzed by NIPT in the study and none of them occurred hemolysis or placed for more than 8 h before treatments. Among these 570 pregnant women, 328 (57.54%) were due to advanced maternal age without other prenatal diagnostic indications and the other 242 (42.46%) were in appropriate age (aged under 35 years at delivery). Among the 242 pregnant women, 80 (14.04%) of them were maternal serological screening high-risk / critical-risk, and 162 (28.42%) of them were abnormal ultrasound diagnosis or undesirable reproductive history. Among the 43 pregnant women with fetal SCAs, 31 were due to advanced maternal age without other prenatal diagnostic indications. The PPV of advanced maternal age group (67.74%) was higher than that of other indication group (45.45%) and serological screening high-risk / critical-risk group (0%), which was consistent with the PPV of 45,X. At the same time, we could figure out that the PPV of 45,X in advanced maternal age group (45.45%) and other prenatal diagnostic indications group (25.00%) were lower than that of other sex chromosomal abnormalities, which was consistent with the previous studies [37, 38], however, we could not draw a credible conclusion because of fewer subjects in other sex chromosomal abnormality group. Of the 43 cases, only one of them was due to high-risk value of serological screening and none due to critical-risk value of serological screening, indicating that although a large number of pregnant women accepted NIPT because of serological screening high-risk / critical-risk, the incidence of fetal SCAs was relatively low.
Conclusions
NIPT based on CPAS could be a potential method for SCAs screening, while this technique needed to be further investigated. It had high false positive rates, especially for 45,X. The pregnant women with fetal SCAs by NIPT, especially those with non-age-related prenatal diagnostic indications, should be advised to accept invasive prenatal karyotype analysis.
Abbreviations
- cfDNA:
-
Cell-free DNA
- cffDNA:
-
Cell-free fetal DNA
- CI:
-
Confidence intervals
- CPAS:
-
Combinatorial Probe-Anchor Synthesis
- DNBs:
-
DNA Nanoballs
- FNR:
-
False negative rate
- FPR:
-
False positive rate
- HRT:
-
Hormone replacement therapy
- ISCN:
-
International System for Human Cytogenetic Nomenclature
- NGS:
-
Next-generation sequencing
- NIPT:
-
Non-invasive prenatal testing
- NPV:
-
Negative predict value
- PPV:
-
Positive predictive value
- RCR:
-
Rolling circle replication
- SCAs:
-
Sex chromosomal aneuploidies
- SEV:
-
Sensitivity
- SPC:
-
Specificity
- TPR:
-
True positive rates
References
Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–7.
Wang T, He QZ, Li HB, Ding J, Wen P, Zhang Q, et al. An optimized method for accurate fetal sex prediction and sex chromosome aneuploidy detection in non-invasive prenatal testing. PLoS One. 2006;11(7):e0159648.
Chandrasekharan S, Minear MA, Hung A, Allyse M. Noninvasive prenatal testing goes global. Sci Transl Med. 2014;6(231):215f–31f.
Allyse M, Minear MA, Berson E, et al. Non-invasive prenatal testing: a review of international implementation and challenges. Int J Womens Health. 2015;7:113–26.
Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401.
Pan X, Zhang C, Li X, Chen S, Ge H, Zhang Y, et al. Non-invasive fetal sex determination by maternal plasma sequencing and applicationin X-linked disorder counseling. J Matern Fetal Neonatal Med. 2014;27(18):1829–33.
Demaliaj E, Cerekja A, Piazze J. Sex chromosome aneuploidies. Aneuploidy in health and disease. In: Storchova Z, editor. Aneuploidy in health and disease. Shanghai, China: In Tech; 2012. p. 123–40.
Warren MP, Chua A. Appropriate use of estrogen replacement therapy in adolescents and young adults with turner syndrome and hypopituitarism in light of the Women’s health initiative. Growth Horm IGF Res. 2006;16:S98–S102.
Abramsky L, Hall S, Levitan J, Marteau TM. What parents are told after prenatal diagnosis of a sex chromosome abnormality: interview and questionnaire study. BMJ. 2001;322(7284):463–6.
Dan S, Wang W, Ren JH, Li YL, Hu H, Xu ZF, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32(13):1225–32.
Patch AM, Nones K, Stephen H, Kazakoff SH, Newell F, Wood S, et al. Germline and somatic variant identification using BGISEQ-500 and HiSeq X ten whole genome sequencing. PLoS One. 2018;13(1):e0190264.
Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327(5961):78–81.
Fehlmann T, Reinheimer S, Geng CY, Su XS, Drmanac S, Alexeev A, et al. cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin Epigenetics. 2016;8:123.
Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105(51):20458–63.
Lau TK, Chen F, Pan X, Pooh RK, Jiang F, Li Y, Jiang H, et al. Noninvasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. J Matern Fetal Neonatal Med. 2012;25(8):1370–4.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–51.
Grati FR, Ferreira JC, Bajaj K. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. Am J Obstet Gynecol. 2014;211(4):365 e1–12.
Yao H, Jiang F, Hu H, Gao Y, Zhu Z, Zhang H, et al. Detection of fetal sex chromosome aneuploidy by massively parallel sequencing of maternal plasma DNA: initial experience in a Chinese hospital. Ultrasound Obstet Gynecol. 2014;44(1):17–24.
Zhang B, Lu BY, Yu B, Zheng FX, Zhou Q, Chen YP, et al. Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood. J Int Med Res. 2017;45(2):621–30.
Benn P. Non-invasive prenatal testing using cell free DNA in maternal plasma: recent developments and future prospects. J Clin Med. 2014;3(2):537–65.
Lau TK, Cheung SW, Lo PS, Pursley AN, Chan MK, Jiang F, et al. Non-invasive prenatal testing for fetal chromosomal abnormalities by low-coverage whole-genome sequencing of maternal plasma DNA: review of 1982 consecutive cases in a single center. Ultrasound Obstet Gynecol. 2014;43(3):254–64.
Maxwell S, Dickinson JE, Murch A, O'Leary P. The potential impact of NIPT as a second-tier screen on the outcomes of high-risk pregnancies with rare chromosomal abnormalities. Aust N Z J Obstet Gynaecol. 2015;55(5):420–6.
Alberry M, Maddocks D, Jones M, Abdel Hadi M, Abdel-Fattah S, Avent N, et al. Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast. Prenat Diagn. 2007;27(5):415–8.
Grati FR, Malvestiti F, Ferreira JC, Bajaj K, Gaetani E, Agrati C, et al. Fetoplacental mosaicism: potential implications for false-positive and false-negative noninvasive prenatal screening results. Genet Med. 2014;16(8):620–4.
Wang Y, Chen Y, Tian F, Zhang J, Song Z, Wu Y, et al. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem. 2014;60(1):251–9.
Mazloom AR, Džakula Ž, Oeth P, Wang H, Jensen T, Tynan J, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequenceng circulating cell-free DNA from maternal plasma. Prenat Diagn. 2013;33(6):591–7.
Jiang F, Ren J, Chen F, Zhou Y, Xie J, Dan S, et al. Noninvasive fetal trisomy (NIFTY) test: an advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Med Genet. 2012;5:57.
Wang JC, Sahoo T, Schonberg S, Kopita KA, Ross L, Patek K, et al. Discordant noninvasive prenatal testing and cytogenetic results: a study of 109 consecutive cases. Genet Med. 2015;17(3):234–6.
Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808.
Choy KW, Kwok KY, Lau ET, et al. Discordant karyotype results among noninvasive prenatal screening positive cases. In: The Shifting Landscape of Genetic Testing: Approaches and Success Stories, Platform Session, Abstract #19, American Society of Human Genetics 2013 Annual Meeting, Boston, MA, USA; 2013.
Meck JM, Dugan EK, Aviram A, et al. Non-invasive prenatal screening: a cytogenetic perspective. In: Oral platform presentations: cytogenetics, abstract #17, American College of Medical Genetics and Genomics 2014 Annual Meeting, Nashville, Tennessee, USA. p. 2014.
Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589–97.
Lutgendorf MA, Stoll KA, Knutzen DM, Foglia LM. Noninvasive prenatal testing:limitations and unanswered questions. Genet Med. 2014;16(4):281–5.
Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013;33(7):667–74.
Lo YM. Fetal DNA in maternal plasma: biology and diagnostic applications. Clin Chem. 2000;46(12):1903–6.
Barrett AN, McDonnell TC, Chan KC, Chitty LS. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin Chem. 2012;58(6):1026–32.
Bianchi DW, Parsa S, Bhatt S, Halks-Miller M, Kurtzman K, Sehnert AJ, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstet Gynecol. 2015;125(2):375–82.
Stone JF, Sandberg AA. Sex chromosome aneuploidy and aging. Mutat Res. 1995;338(1–6):107–13.
Acknowledgements
We would like to thank our patients for agreeing to donate their personal data to our study and have these been published. We are grateful to Dr. Jiong Gao and Dr. Ya Gao (BGI Genomics, BGI-shenzhen 518083, China) for analyzing the data and modifying the article.
Funding
This work is supported by the National Key Research and Development Program of China (2016YFC1000703), the Medical Scientific Research Foundation of Zhejiang Province (2017KY427; 2018KY809) and the Science and Technology Department Foundation of Zhejiang Province (2017C3303).
Availability of data and materials
The data supporting the conclusions of this article is included within the article.
Author information
Authors and Affiliations
Contributions
All authors have participated in the study and manuscript preparation. LHG and LY performed all the molecular genetic analysis and drafted the manuscript, ZH and LYQ performed karyotype analysis, QYQ and CM participated in conceiving the work and revising the manuscript, SYX collected all clininal data, YK and YYM performed the NIPT. LB and WLY performed the statistical analysis and helped to draft the manuscript. HYZ and HJJ took a plan for the research. DMY designed the work, drafted and revised the manuscript. XJY revised the manuscript and improved the written English. All authors have approved the final article.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Hospital Ethics Committee of the Women’s Hospital School of Medicine, Zhejiang University. Each patient signed informed consent prior to study enrollment.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, H., Lei, Y., Zhu, H. et al. The application of NIPT using combinatorial probe-anchor synthesis to identify sex chromosomal aneuploidies (SCAs) in a cohort of 570 pregnancies. Mol Cytogenet 11, 59 (2018). https://doi.org/10.1186/s13039-018-0407-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-018-0407-z