Abstract
Background
Homozygous mutations and deletions of the microcephalin gene (MCPH1; OMIM *607117) have been identified as a cause of autosomal recessive primary microcephaly and intellectual disability (MIM #251200). Previous studies in families of Asian descent suggest that the severity of the phenotype may vary based on the extent of the genomic alteration. We report chromosome microarray (CMA) findings and the first described family study of a patient with primary microcephaly in a consanguineous Hispanic family.
Case presentation
The proband, a boy born at full-term to consanguineous parents from Mexico, presented at 35 months of age with microcephaly, abnormal brain MRI findings, underdeveloped right lung, almond-shaped eyes, epicanthal folds, bilateral esotropia, low hairline, large ears, smooth philtrum, thin upper lip, and developmental delay. MRI of the brain showed a small dermoid or lipoma (without mass effect) within the interpeduncular cistern and prominent arachnoid granulation. The underdeveloped right lung was managed with long-acting inhaled corticosteroids. Otherwise the proband did not have any other significant medical history. The proband had 2 older brothers, ages 14 and 16, from the same consanguineous parents. The 14-year-old brother had a phenotype similar to that of the proband, while both parents and the oldest brother did not have the same phenotypic findings as the proband. The SNP-based CMA analysis of the proband detected a homozygous 250-kb microdeletion at 8p23.2p23.1, extending from 6,061,169 to 6,310,738 bp [hg19]. This genomic alteration encompasses the first 8 exons of MCPH1. Follow-up studies detected the same homozygous deletion in the affected brother, segregating with microcephaly and intellectual disability. Regions of homozygosity (ROHs) were also observed in the affected brother. Since ROHs are associated with an increased risk for recessive disorders, presence of ROH may also contribute to the phenotype of the affected brothers. The parents were both hemizygous for the deletion.
Conclusion
Here we report a homozygous deletion of multiple exons of the MCPH1 gene that was associated with primary microcephaly and intellectual disability in a Hispanic family. In the context of previous studies, our results support the idea that deletions involving multiple exons cause a more severe phenotype than point mutations.
Similar content being viewed by others
Background
Primary microcephaly (MCPH) is a genetically heterogeneous disorder, predominately showing an autosomal recessive mode of inheritance. MCPH is a developmental defect of the brain characterized by a congenital small cranium with a reduced occipito-frontal head circumference (OFC) of more than 2 standard deviations (SD) below the mean for age, sex, and ethnicity (severe microcephaly OFC < 3 SD). MCPH is also characterized by mild to moderate intellectual disability and mild seizures, simplified gyral pattern, periventricular neuronal heterotopias, polymicrogyria, speech delay, hyperactivity and attention deficit, aggressiveness, focal or generalized seizures, and delay of developmental milestones and pyramidal signs [1, 2].
Mutations in 18 genes are known to cause MCPH; MCPH1 (*607117) was the first gene to be identified [3, 4]. Whole exome (WES) or the whole genome (WGS) studies revealed an additional 17 OMIM genes associated with MCPH including WDR62 (*613583), CDK5RAP2 (*608201), CASC5 (*609173), ASPM (*605481), CENPJ (*609279), STIL (*181590), CEP135 (*611423), CEP152 (*613529), ZNF335 (*610827), PHC1 (*602978), CDK6 (*603368), CENPE (*117143), CENPF (*600235), PLK4 (*605031), TUBGCP6 (*610053), CEP63 (*614724), NDE1 (*609449) [5].
MCPH caused by MCPH1 mutation presents with congenital microcephaly, intellectual disability, and a head circumference that is reduced to a greater degree than height [6].
The MCPH1/microcephalin gene (*607117) is located at chromosome 8p23 and has a genomic size of 241,905 bp. The open reading frame is 8032 bp and consists of 14 exons that encode 835 amino acids; 3 isoforms have been reported so far [4]. Microcephalin, the encoded protein, is implicated in chromosome condensation and cellular responses that are induced by DNA damage [7]. This protein is thought to have a role in G2/M checkpoint arrest via maintenance of inhibitory phosphorylation of cyclin-dependent kinase 1 [7,8,9]. Loss of the microcephalin protein thus triggers early mitotic entry of neuroprogenitor cells leading to the inability to maintain brain size [7,8,9].
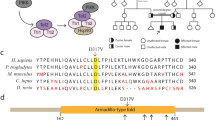
Two types of mutations in MCPH1 have been reported in the literature. A missense mutation was identified in patients with a less severe cellular phenotype and mild microcephaly [6, 10, 11]. Deletions in MCPH1 have also been reported. A deletion of the first 6 exons of the gene was identified in an Iranian family with intellectual disability and mild microcephaly; premature chromosome condensation in at least 10% to 15% of cells was also reported for this family [11]. In addition, a deletion of the first 11 exons was identified in an Asian-Indian patient [12].
Here we present a consanguineous Hispanic family with primary microcephaly and intellectual disabilities associated with MCPH1 deletions.
Case presentation
The proband presented at 35 months of age with microcephaly (44 cm head circumference, which is more than 2 SD below the third percentile for age), dysmorphic facial features (almond shaped eyes, epicanthal folds, bilateral esotropia, low hairline, large ears, smooth philtrum, and thin upper lip), and intellectual disability or developmental delay.
The proband’s MRI of the brain showed a small dermoid or lipoma (without mass effect) within the interpeduncular cistern and prominent (1.2 × 0.4 cm) arachnoid granulation (Fig. 1). The proband also had an underdeveloped right lung, which was managed with long-acting inhaled corticosteroids. Otherwise the proband did not have any other significant medical history. The proband was born via full-term vaginal delivery at 2720 g. The 34-year old mother was G4P3A1L3 and had no known maternal complications or infections. The proband had no prenatal, perinatal, or postnatal complications, and he was discharged on day 2 of life. He did not walk until he was 13 months and did not speak words until 2 years old, so he received physical and speech therapy for diagnosis of developmental delay. At 48 months, <25% of his speech was intelligible to strangers, and he did not form sentences.
The proband had two brothers who were 14 and 16 years old and from the same consanguineous parents (Fig. 2). One of the brothers (V.6) presented with a similar phenotype as the proband (V.8), while both parents were phenotypically normal (IV.6 and IV.7). The pedigree indicated consanguinity in offspring who had microcephaly, intellectual disabilities, ptosis, hearing loss, and short stature. Hypertension, diabetes, high cholesterol, arthritis, stroke, and osteoporosis were also present in these offspring.
The consanguineous parents of the proband had normal head circumferences for their ages (57 cm for the father, 57.5 cm for the mother). They did not finish high school and stated it was due to marriage at young age. Their country of origin was Mexico. There was no history of seizures or cardiac, renal, skeletal, or metabolic disease noted in the proband or any of his first degree relatives. In order to determine the cause of the patient’s abnormality, CMA was ordered for both parents.
Methods
Genomic DNA was extracted from whole blood using the Gentra Puregene kit (Qiagen-Sciences, Maryland, USA). Microdeletion/microduplication screening was performed for the proband, his parents, and available brothers using a SNP-array platform (CytoScan HD; Affymetrix, Santa Clara, CA), following the manufacturer’s instruction. The CytoScan HD array has 2.67 million probes, including 1.9 million copy number probes and 0.75 million SNP probes. Array data were analyzed using the Chromosome Analysis Suite (ChAS) (Affymetrix, Inc.) software v2.0. FISH analysis using BlueGnome probes RP11-11I1 (Illumina, San Diego, CA, USA) for the deleted region 8p23.2p23.1 was performed on metaphase cells according to the manufacturer’s protocol. Subsequently, 50 cells were examined carefully.
Results and discussion
The SNP-microarray analysis of the proband’s DNA identified a homozygous microdeletion of 250 kb at 8p23.2p23.1 (Fig. 3). The deletion extended from 6,061,169 to 6,310,738 bp (UCSC genome Browser; http://genome.ucsc.edu/; hg19 release). This genomic alteration indicated a homozygous loss (zero copy) of the first 8 exons of the microcephalin gene (MCPH1; OMIM #607117).
Chromosome microarray analysis. Chromosome 8 with deletion at 8p23.2p23.1 is shown along the right side of the image. SNP-array results show the copy number state (black pedigree labels), weighted log2 ratio (green pedigree labels), and allele peaks (red pedigree labels) for proband (V.8), his brother (V.6), his mother (IV.6), his father (IV.7), and the MCPH1 gene at the deleted region
The homozygous deletion detected in our proband was also observed in one of his brothers and segregated with microcephaly and intellectual disability (Fig. 3). The parents were both found to be hemizygous for this deletion (Fig. 3) and admitted to consanguinity, which supported the autosomal recessive nature of this pathogenic deletion for microcephaly and intellectual disability. FISH testing confirmed homozygous deletion of the MCPH1 gene in the proband and hemizygous deletion of the same gene in his mother and father (Fig. 4a, b). The older, unaffected brother was not available for testing.
The clinical and molecular features of the affected siblings presented herein are similar to those of patients reported in the literature to also have MCPH1 deletions (Table 1) [11, 12]. Garshasbi et al. reported 6 individuals (4 males, 2 females) of an Iranian family who had primary microencaphly had a 150-200 kb deletion (8p22.2p23.2), which spanned 25 kb of the promoter sequence of MCPH1 and exons 1-6 [11]. Microcephaly in these individuals was borderline to moderate and mental retardation was mild to moderate [11]. A single case report of primary microencaphly caused by a microdeletion exists of a male Asian Indian 2 month-old baby [12]. For this subject, the deletion (8p23.1(6197,889-6295,040)×1) involved exons 1-11 of MCPH1 [12]. From these few reports it appears that greater genomic alteration of the MCPH1 gene may be indicative of more significant clinical phenotypes, since studies of patients with point mutations in MCPH1 suggest milder phenotypes [13].
In addition to homozygous copy number loss, many copy neutral regions of homozygosity (ROHs) were observed in both the patient (Fig. 5a) and his brother (Fig. 5b). ROHs that span >5 Mb totaled approximately 148 Mb across the genome. The implications of ROHs are unclear at present. In theory, when the degree of homozygosity is significant, there is an increased risk for recessive Mendelian disorders. Since the clinical features of our patient and the previously reported cases with exonic deletions are similar, the ROHs found in our patient are likely to have no or minimum impact on the patient’s phenotype.
Conclusions
Here we report a homozygous deletion of multiple exons of the MCPH1 gene that was associated with primary microcephaly and intellectual disability; to our knowledge, this is the first such report in a Hispanic family. In the context of previous studies, our results support the idea that deletions involving multiple exons cause a more severe phenotype than point mutations.
Abbreviations
- CMA:
-
Chromosomal microarray analysis
- MCPH1:
-
Microcephalin gene
- Oligo-SNP array:
-
Oligonucleotide-single nucleotide polymorphism array
References
Passemard S, Titomanlio L, Elmaleh M, Afenjar A, Alessandri JL, Andria G, et al. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology. 2009;73:962–9.
Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, et al. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90:363–83.
Jackson AP, McHale DP, Campbell DA, Jafri H, Rashid Y, Mannan J, et al. Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet. 1998;63:541–6.
Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–42.
Morris-Rosendahl DJ, Kaindl AM. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH). Mol Cell Probes. 2015;29:271–81.
Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 2005;76:717–28.
Barbelanne M, Tsang WY. Molecular and cellular basis of autosomal recessive primary microcephaly. Biomed Res Int. 2014;2014:547986.
Cox J, Jackson AP, Bond J, Woods CG. What primary microcephaly can tell us about brain growth? Trends Mol Med. 2006;12:358–66.
Gruber R, Zho Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–34.
Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–6.
Gharshasbi M, Motazacker MM, Kahrizi K, Behjati F, Abedini SS, Nieh SE, et al. SNP array-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Hum Genet. 2006;118:708–15.
Pfau RB, Thrush DL, Hamelberg E, Bartholomew D, Botes S, Pastore M, et al. MCPH1 deletion in a newborn with severe microcephaly and premature chromosome condensation. Eur J Med Genet. 2013;56:609–13.
Darvish H, Esmaeeli-Nieh S, Monajemi GB, Mohseni M, Ghasemi-Firouzabadi S, Abedini SS, et al. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J Med Genet. 2010;47:823–8.
Acknowledgements
The authors acknowledge that Andrew Hellman, PhD of Scientific Publication who offered valuable assistance in manuscript revisions.
Funding
Not applicable.
Availability of data and materials
The raw data and materials used for this study have been de-identified and were freely available to any scientist wishing to use them without breaching participant confidentiality.
Author information
Authors and Affiliations
Contributions
MH designed and conducted the project, drafted and finalized the manuscript. The rest of the authors provided input to the project, made critical comments on the drafted manuscript, gathered clinical information. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Authors’ information
Morteza Hemmat - the first and corresponding author. The clinical information was provided by Melissa J Rumple. The rest of the authors helped to draft the manuscript All authors read and approved the final manuscript.
Ethics approval and consent to participate
Consent form was signed by the patient’s parent and it is available upon request.
Consent for publication
Consent forms for publication and pictures was signed by the patient’s parent and it is available upon request.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hemmat, M., Rumple, M.J., Mahon, L.W. et al. CMA analysis identifies homozygous deletion of MCPH1 in 2 brothers with primary Microcephaly-1. Mol Cytogenet 10, 33 (2017). https://doi.org/10.1186/s13039-017-0334-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-017-0334-4