Abstract
Background
Porcine sapovirus, belonging to the family Caliciviridae, is an enteric virus that is widespread in the swine industry worldwide. A total of 14 sapovirus genogroups have been suggested and the most commonly found genogroup in swine is genogroup III (GIII). The goal of the present experiment was to examine the presence of sapovirus in 51 naturally infected pigs at two different time points. The pigs were kept under experimental conditions after weaning. Previous studies on sapovirus have primarily been of a cross sectional nature, typically prevalence studies performed on farms and abattoirs. In the present study, faecal samples, collected from each pig at 5½ weeks and 15–18 weeks of age, were analysed for sapovirus by reverse transciptase polymerase chain reaction and positive findings were genotyped by sequencing.
Results
At 5½ weeks of age, sapovirus was detected in the majority of the pigs. Sequencing revealed four different strains in the 5½ week olds—belonging to genogroups GIII and GVII. Ten to 13 weeks later, the virus was no longer detectable from stools of infected pigs. However, at this time point 13 pigs were infected with another GIII sapovirus strain not previously detected in the pigs studied. This GIII strain was only found in pigs that, in the initial samples, were virus-negative or positive for GVII.
Conclusions
At 5 weeks of age 74 % of the pigs were infected with sapovirus. At 15–18 weeks of age all pigs had cleared their initial infection, but a new sapovirus GIII strain was detected in 25 % of the pigs. None of the pigs initially infected with the first GIII strain were reinfected with this new GIII strain, which may indicate the presence of a genogroup-specific immunity.
Similar content being viewed by others
Background
Sapovirus is a genus of the family Caliciviridae. Human sapovirus is known to cause acute gastroenteritis in children [1]. In swine, experimental infection with porcine sapovirus has also been associated with intestinal disease [2, 3]. In pig herd studies from several countries worldwide, sapoviruses have been reported to be present both in pigs with and without diarrhoea [4–12] leaving uncertainty about the magnitude of their role in enteric disease in pigs. Prevalence studies have shown that the virus is widespread and occurs worldwide in the pig industry, however, the reported prevalence varies from 3 to 67 % [5, 7, 10, 12–14]. Though the prevalence of sapovirus in pigs varies between countries, the highest prevalence is generally found in post-weaning pigs [7].
Sapoviruses are 35 nm sized non-enveloped viruses with a single-stranded positive-sense RNA genome of approximately 7.3 kb in length. The genome is composed of two open reading frames (ORF) where ORF1 encodes the non-structural proteins and the major capsid protein, VP1, whereas ORF2 encodes the minor structural protein VP2 [15, 16]. Based on the complete capsid gene sequences sapoviruses are divided into seven genogroups (GI-GVII) [15]. Pigs have frequently been reported to primarily host GIII sapoviruses [5, 8, 10, 12–14], but other studies have revealed a more extensive genetic heterogeneity among porcine sapoviruses. Thus a wider range of sapovirus genogroups including several proposed genogroups (GVI to GXI), have been detected in swine faeces [7, 9, 15, 17–21]. Based on the sequence of the RNA polymerase region, some of these swine sapovirus genogroups are more closely related to the human genogroups than to the GIII sapoviruses initially reported in swine.
The previous studies of sapoviruses in pigs have primarily been of a cross-sectional nature thus it is not known how sapovirus infections develop over time.
In the present study, we tested 51 post-weaning pigs twice for the presence of sapovirus. The pigs were naturally infected but kept under experimental conditions. Positive virus findings were characterized by sequencing, and the repeated sampling allowed comparison of the infections in each individual pig at two points in time.
Methods
Experimental set up
Two groups, A and B, comprised of 26 and 25 pigs respectively, were studied. The pigs were part of two identical vaccine trials against Mycoplasma hyosynoviae—a bacterium that causes arthritis and has no impact on the gastrointestinal system in infected swine. Therefore it was appropriate to conduct a parallel study of sapovirus occurrence in these animals. The two trials were run eight months apart in 2008 (A) and 2009 (B). The pigs were cross-breeds (Danish Landrace, Yorkshire, Duroc) and represented six age matched litters that originated from a Danish specific-pathogen-free swine herd. The herd was declared free from Mycoplasma hyopneumoniae, toxigenic Pasteurella multocida, Bracyspira hyodysenteria, Porcine reproductive and respiratory syndrome virus, Haematopinus suis, Sarcoptes scabiei and all Actinobacillus pleuropneumoniae serotypes except from serotype 12. Testing the herd for the presence of sapovirus was not performed prior to the experiment. The pigs were transferred to the experimental facilities after weaning at 3½–4 weeks of age and were kept there during the sampling period. As a standard procedure, the pens of the experimental facilities had been disinfected with 1 % Wofasteril 050 (BiOsense, Tønder, DK) (Group A) or 1 % Virkon S (Antec International, Sudbury, UK) (Group B) before use. In Group A three litters were distributed in four pens with 9 pigs (litter 1), 7 (litter 2), 5 and 5 pigs (litter 3) in each pen, respectively, without mixing the litters. In Group B three litters were distributed, without mixing the litters, in three pens of 9, 5 and 11 pigs, respectively. However, in Group B, at 7 weeks of age, a more even distribution of pigs in the three pens was desired and four pigs were transferred from the pen with the 11-pig litter to the pen with the smallest litter; thus obtaining 9, 9 and 7 pigs in the pens. Furthermore, in Group B, as part of a modification of the M. hyosynoviae infection model, at 13 weeks of age, half of the pigs in each pen were transferred to another of the three pens, thus mixing the litters within the second trial of the study.
The pens had concrete floors and bedding consisting of sawdust and straw. The pigs were fed a factory-made pelleted standard swine feed. No antimicrobials were added to the feed during the experiment. At weaning, all pigs had a normal body condition and did not show any clinical signs of disease. The study was approved by the Danish Animal Experiments Inspectorate (approval no. 2006/561-1106, protocol no. 70), and performed according to Danish legislation.
When the pigs were 5½ weeks old, faecal samples were collected manually from the rectum. Disposable plastic gloves were used for each sample and disposed between each sampling/pig. When the pigs were 15–16½ weeks old (Group A) or 16½–18 weeks old (Group B) follow-up faecal samples were obtained. All faecal samples were stored in sterile tubes at 4 °C until processing.
Virus detection and sequencing
Faecal suspensions in 10 % phosphate buffered saline (PBS) were prepared, and after centrifuging RNA was extracted by using a total nucleic acids kit on a MagNa pure LC robot (Roche Diagnostics A/S, Hvidovre, Denmark). For the detection of sapovirus, two different reverse transciptase polymerase chain reaction (RT-PCR) tests, targeting the polymerase region, were used in all samples. In the first RT-PCR the primers used were p290/p289 [22] and in the second RT-PCR the same primers, in combination with four other p290/p289 derived primer pairs [23], were used. The PCR products were all 286 base pairs long exclusive of the primers. For RT-PCR, a one-step RT-PCR kit (QIAGEN Nordic, Copenhagen, Denmark) was used with the following reverse transcription and cycling conditions: a transcription step at 50 °C for 30 min, followed by 95 °C for 15 min and 40 cycles at 94 °C for 30 s, 49 °C for 30 s and 72 °C for 30 s with a final extension step at 72 °C for 10 min. The PCR products were sequenced in both directions on an ABI 3100 instrument using a BigDye kit v 1.1, (Applied Biosystems, Nærum, Denmark) and quality assessed and assembled using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium). The sapovirus sequences obtained in the present study were deposited in GenBank with the following accession numbers: FJ947001, FJ947002, GU173811, GU173812 and GU320723.
The phylogenetic tree was constructed using the UPGMA clustering method (MEGA 4).
Results
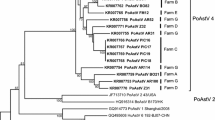
The number of animals that tested positive for sapoviruses in relation to group, litter, sampling time, and genogroups is presented in Table 1. Sequencing of the PCR products showed that the sapoviruses belonged to two genogroups GIII and GVII (Fig. 1). Within GIII, four different strains, designated A–D, were detected. These strains differed 6–27 % in the nucleotide sequences and 4–10 % on amino acid level in the polymerase region (286 base pairs). However, all samples of the same strain (A: n = 15, B: n = 3, C: n = 5, D: n = 13) had 100 % identical nucleotide sequences in the polymerase region studied (286 base pairs).
Phylogenetic tree based on 286 base pairs long RT-PCR sequences showing the sapovirus strains obtained. Five sapovirus strains were obtained in this study (shown in bold). The prototype strains PEC-Cowden (AF18276) and Sapporo (U65427) are shown in italics. The sequences designated with GenBank accession numbers are from a previous study [7]. The bootstrap values (500 replicates) are plotted at selected internal branch nodes. The tree is rooted with the out-group Sapporo (U65427). The scale bar represents nucleotide substitutions per site
In Group A, all follow-up samples were sapovirus negative, but in Group B, half of the pigs (n = 13) were sapovirus positive at the end of the experiment. Sequencing showed that these 13 pigs had been infected with a new sapovirus strain (GIII strain D) and that the sequences were 100 % identical. None of the two strains detected initially in Group B were found in follow-up samples. The GIII strain D was only detected in pigs that had been either sapovirus negative (n = 5) or GVII positive (n = 8) at 5½ weeks of age, and not in any of the five pigs that had been positive for GIII strain C at the first sampling.
Discussion
Sapovirus infections are common in swine worldwide [7, 14, 18, 24]. The virus has been detected in pigs of all age groups, but is most frequently reported in post-weaning pigs [7, 11–14, 17, 24–26]. In the present study sapovirus was detected in 82 % of the pigs in Group A and 68 % of the pigs in Group B when sampled at 5½ weeks of age. The pigs originated from six different litters from one specific-pathogen-free herd, and they were healthy and in good condition at the initiation of the experiment.
The sapovirus strains detected in the pigs at 5½ weeks of age (GIII strain A, B, and C and GVII) could not be recovered in the second sampling when the pigs were 15–18 weeks old. However, we found that 13 of the 51 pigs excreted another sapovirus strain, GIII strain D, in the second sampling. Thus, in this experiment, more pigs were sapovirus-positive at 5½ weeks of age compared to 15–18 weeks. This indicates that pigs are transient shedders of sapovirus and confirms the findings of others [7, 14, 19] that sapovirus infections are highly prevalent in post-weaning pigs. However, Wang et al. [14] who investigated 621 faecal samples from American herds and abattoirs also found a high prevalence of sapovirus infections in finishers and sows, whereas they seemed to be less prevalent in nursing pigs. In Korea, Song et al. [19] investigated 567 faecal samples and found 37 sapovirus isolates of which 59.5 % derived from post-weaners, whereas 32.4 % were from nursing pigs and 8.1 % from growing pigs. It is possible that the more widespread detection of sapovirus across age-groups relates to a number of different sapovirus strains circulating within the farms and that older pigs will typically be sapovirus positive due to a reinfection. This was also observed in Group B in our study, where half of the pigs were sapovirus positive at 16½–18 weeks of age due to infection with a sapovirus of a new genogroup compared to earlier findings in the same pigs.
An interesting observation was that none of the pigs infected with a GIII virus at 5½ weeks of age, experienced any reinfection. The sapovirus strain causing reinfections in 13 pigs was also GIII (Table 1), but differed 7 % in the amino sequence in the polymerase region from the GIII strain causing the initial infection. This finding could indicate the existence of a genogroup-specific immunity, of at least a short duration. Furthermore, it also indicates the lack of cross-neutralization between the two different genogroups GIII and GVII. Such homotypic immunity has been described to be present in humans when infected with norovirus—another member of the Caliciviridae family [27, 28]. However, specific identification of a homotypic immunity in pigs infected with sapovirus and the duration of such immunity, would need further studies.
To improve the possibility of finding more than one genotype of sapovirus, two different RT-PCR tests, targeting the polymerase region, were used for the detection of sapoviruses in all samples. However, both test systems detected the same viruses in this setting. In another study, we detected different sapovirus genogroups and strains when testing faecal samples from swine with more than one RT-PCR assay. This was interpreted as double infections [7]. In the present study, five different sapovirus strains were detected, and the same virus strain was found in as many as 15 pigs. As sapoviruses are single-stranded RNA viruses with a high mutation rate, the finding of identical sequences indicates a common point source of infection. This source could be the housing environment or other animals e.g. the sow. Strict isolation rules were applied in the stable with respect to outer surroundings, but not between the pens within the experiment. Therefore spread of infection from animals in one pen to another may have occurred by vehicle transmission by e.g. footwear, personnel or tools. For unknown reasons we found an additional strain (GIII strain D), not earlier detected in this study, in the second sampling in Group B. This was unexpected since we would expect that all sapovirus sequences in the study would have originated from the herd of origin. However, sapoviruses can survive outside the host for considerable time and thorough disinfection with special disinfectants is needed to inactivate the virus [15]. The litters in Group B were mixed twice before the second sampling, a fact that might have affected the results as the pigs could have been in contact with new surroundings and viruses during the transfers. The introduction of the GIII strain D might be explained by either, insufficient disinfection, introduction via the personnel or introduction via bedding material/feed. The reason still remains unidentified.
Conclusions
We found that the vast majority of the 5½ week old pigs carried sapovirus whereas the infection, though present, was less frequent in the pigs in the second sampling at 15–18 weeks of age. Pigs shedding sapovirus at the second sampling all had an infection with a different genotype than at the first sampling, indicating that they had cleared the initial infection. Thus, reinfection occurs, but genogroup-specific immunity might exist for sapovirus, at least of a short duration, such as it has been described for other members of the Caliciviridae family in humans. If this is the case, it would be relevant to investigate the duration of such immunity and in addition, which parts of the immune system may be providing such immunity.
Abbreviations
- GIII :
-
sapovirus genogroup III
- GVI :
-
sapovirus genogroup GVI
- GVII :
-
sapovirus genogroup VII
- GXI :
-
sapovirus genogroup GXI
- ORF:
-
open reading frame
- PBS:
-
phosphate buffered saline
- PCR:
-
polymerase chain reaction
- RNA:
-
ribonucleic acid
- RT-PCR:
-
reverse transciptase polymerase chain reaction
- VP1:
-
major capsid protein of sapovirus
- VP2:
-
minor structural protein of sapovirus
References
Schlenker C, Surawicz CM. Emerging infections of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2009;23:89–99.
Flynn WT, Saif LJ, Moorhead PD. Pathogenesis of porcine enteric calicivirus-like virus in four-day-old gnotobiotic pigs. Am J Vet Res. 1988;49:819–25.
Guo M, Hayes J, Cho KO, Parwani AV, Lucas LM, Saif LJ. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J Virol. 2001;75:9239–51.
Collins PJ, Martella V, Buonavoglia C, O’Shea H. Detection and characterization of porcine sapoviruses from asymptomatic animals in Irish farms. Vet Microbiol. 2009;139:176–82.
Keum HO, Moon HJ, Park SJ, Kim HK, Rho SM, Park BK. Porcine noroviruses and sapoviruses on Korean swine farms. Arch Virol. 2009;154:1765–74.
Mauroy A, Scipioni A, Mathijs E, Miry C, Ziant D, Thys C, Thiry E. Noroviruses and sapoviruses in pigs in Belgium. Arch Virol. 2008;153:1927–31.
Reuter G, Zimsek-Mijovski J, Poljsak-Prijatelj M, Di Bartolo I, Ruggeri FM, Kantala T, et al. Incidence, diversity and molecular epidemiology of sapoviruses in swine across Europe. J Clin Microbiol. 2010;48:363–8.
Zhang W, Shen Q, Hua X, Cui L, Liu J, Yang S. The first Chinese porcine sapovirus strain that contributed to an outbreak of gastroenteritis in piglets. J Virol. 2008;82:8239–40.
Cunha JB, de Mendonça MC, Miagostovich MP, Leite JP. Genetic diversity of porcine enteric caliciviruses in pigs raised in Rio de Janeiro State, Brazil. Arch Virol. 2010;155:1301–5.
Liu GH, Li RC, Huang ZB, Yang J, Xiao CT, Li J, et al. RT-PCR test for detecting porcine sapovirus in weanling piglets in Hunan Province, China. Trop Anim Health Prod. 2012;44:1335–9.
Martínez MA, Alcalá AC, Carruyo G, Botero L, Liprandi F, Ludert JE. Molecular detection of porcine enteric caliciviruses in Venezuelan farms. Vet Microbiol. 2006;116:77–84.
Dufkova L, Scigalkova I, Moutelikova R, Malenovska H, Prodelalova J. Genetic diversity of porcine sapoviruses, kobuviruses, and astroviruses in asymptomatic pigs: an emerging new sapovirus GIII genotype. Arch Virol. 2013;158:549–58.
Yu JN, Kim MY, Kim DG, Kim SE, Lee JB, Park SY, et al. Prevalence of hepatitis E virus and sapovirus in post-weaning pigs and identification of their genetic diversity. Arch Virol. 2008;153:739–42.
Wang QH, Souza M, Funk JA, Zhang W, Saif LJ. Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridazation assays. J Clin Microbiol. 2006;44:2057–62.
Di Bartolo I, Tofani S, Angeloni G, Ponterio E, Ostanello F, Ruggeri FM. Detection and characterization of porcine caliciviruses in Italy. Arch Virol. 2014;159:2479–84.
Bank-Wolf BR, König M, Thiel HJ. Zoonotic aspects of infections with noroviruses and sapoviruses. Vet Microbiol. 2010;140:204–12.
Wang QH, Han MG, Funk JA, Bowman G, Janies DA, Saif LJ. Genetic diversity and recombination of porcine sapoviruses. J Clin Microbiol. 2005;43:5963–72.
L’Homme Y, Sansregret R, Plante-Fortier E, Lamontagne AM, Lacroix G, Ouardani M, et al. Genetic diversity of porcine Norovirus and Sapovirus: Canada, 2005–2007. Arch Virol. 2009;154:581–93.
Song YJ, Yu JN, Nam HM, Bak HR, Lee JB, Park SY, et al. Identification of genetic diversity of porcine Norovirus and Sapovirus in Korea. Virus Genes. 2011;42:394–401.
Scheuer KA, Oka T, Hoet AE, Gebreyes WA, Molla BZ, Saif LJ, Wang Q. Prevalence of porcine noroviruses, molecular characterization of emerging porcine sapoviruses from finisher swine in the United States, and unified classification scheme for sapoviruses. J Clin Microbiol. 2013;51:2344–53.
Martella V, Lorusso E, Banyai K, Decaro N, Corrente M, Elia G, et al. Identification of a porcine calicivirus related genetically to human sapoviruses. J Clin Microbiol. 2008;46:1907–13.
Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J Virol Methods. 1999;83:145–54.
Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, et al. Genetic diversity among sapoviruses. Arch Virol. 2004;149:1309–23.
Wang QH, Costantini V, Saif LJ. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine. 2007;25:5453–66.
Barry AF, Alfieri AF, Alfieri AA. High genetic diversity in RdRp gene of Brazilian porcine sapovirus strains. Vet Microbiol. 2008;131:185–91.
Shen Q, Zhang W, Yang S, Chen Y, Ning H, Shan T, et al. Molecular detection and prevalence of porcine caliciviruses in eastern China from 2008 to 2009. Arch Virol. 2009;154:1625–30.
Johnson PC, Mathewson JJ, DuPont HL, Greenberg HB. Multiple-challenge study of host susceptibility to Norwalk gastroenteritis in US adults. J Infect Dis. 1990;161:18–21.
Bull RA, White PA. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 2011;19:233–40.
Authors’ contributions
KTL performed the design of the study, planned the animal experiments, performed the animal experiments, collected the faecal samples. Participated in analysis and interpretation of the results. Drafted and submitted the manuscript. MSH participated in the design of the study and planning of the animal experiments, performed the animal experiments, collected the faecal samples. Helped with interpreting the results and participated in drafting the manuscript. CKJ carried out the RT-PCR, sequencing and the sequence alignments, including the analysis of results. BB participated in the planning of the experiment and the interpretation of the results obtained. Participated in the design and coordination of the study. Performed analysis of the results. Participated in drafting the manuscript. GJ participated in planning the experiment, helped with interpreting the results and participated in drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was financed by Danish Pig Production and The Danish AgriFish Agency—Ministry of Food, Agriculture and Fisheries of Denmark.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lauritsen, K.T., Hansen, M.S., Johnsen, C.K. et al. Repeated examination of natural sapovirus infections in pig litters raised under experimental conditions. Acta Vet Scand 57, 60 (2015). https://doi.org/10.1186/s13028-015-0146-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13028-015-0146-7