Abstract
Background
Quantitative real-time reverse transcription PCR (qRT-PCR) requires a stable internal control to avoid misinterpretation of data or errors for gene expression normalization. However, there are still no validated reference genes for stable internal control in Poria cocos (Schw.) Wolf (Fuling). This study aims to validate the reference genes of P. cocos.
Methods
This study firstly collected the 14 candidate reference genes by BLASTP from the genome of P. cocos for qRT-PCR analysis to determine the expression levels of 14 housekeeping genes (GAPDH, MAPK, β-Act, RPB2, RPB1-1, RPB1-2, his3-1, his3-2, APT, SAMDC, RP, β-Tub, EIF, and CYP) under different temperatures and in response to different plant hormones (indole-3-acetic acid, abscisic acid, 6-benzylaminopurine, methyl jasmonate, and gibberellic acid), and the threshold cycle (Ct) values. The results were analyzed by four programs (i.e., geNorm, NormFinder, BestKeeper, and RefFinder) for evaluating the candidate reference genes.
Results
SAMDC, his3-2, RP, RPB2, and his3-1 were recommended as reference genes for treating P. cocos with indole-3-acetic acid, abscisic acid, 6-benzylaminopurine, methyl jasmonate, and gibberellic acid, respectively. Under different temperatures RPB2 was the most stable reference gene. CYP was the most stable gene for all 90 samples by RefFinder.
Conclusion
SAMDC, his3-2, RP, RPB2, and his3-1 were evaluated to be suitable reference genes for P. cocos following different treatments. RPB2 was the most stable reference gene under different temperatures and CYP was the most stable gene in the mycelia under all six evaluated conditions.
Similar content being viewed by others
Background
Quantitative real-time reverse transcription PCR (qRT-PCR) is used for determining the abundance of mRNAs in molecular biology studies. Suitable reference genes are necessary to ensure accuracy and to avoid bias. Typically, reference genes are housekeeping genes necessary for cellular metabolism. The genes for cyclophilin (CYP), tubulin, ubiquitin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin, 18S ribosomal RNA, 28S ribosomal RNA, and albumin are among the most frequently used reference genes [1].
However, the expression levels of reference genes may not be stable in different species [2], different tissues [3], or even identical cells under different culture conditions [4]. For example, the biosynthesis of triterpenes was induced by methyl jasmonate (MeJA) in Ganoderma lucidum (Leyss. ex Fr.) P. Karst (Lingzhi) [5, 6]. However, the stability of fungal reference genes in the presence of plant hormones has not been properly evaluated by the gene expression levels of enzymes involved in the triterpene biosynthesis pathway.
Little research has been conducted on reference genes in fungi. In Hemileia vastatrix Berk. and Br. (Toubaoxiujun), the cytochrome b, 40S ribosomal protein and Hv00099 genes have been selected as reference genes in vitro; however, the 40S ribosomal protein, GAPDH, and Hv00099 genes were the most stable genes in planta [7]. In Hypocrea jecorina Berk. and Br. (Hongherouzuojun), the gene encoding a GTPase was recommended as a reference gene [8]. Reference genes for qRT-PCR under different culture conditions and at different developmental stages in G. lucidum were reported [9].
Poria cocos (Schw.) Wolf (Fuling) is medicinal fungi and nutrition food widely distributes in East Asia, particularly in China, North America, Africa, and Australia [10, 11]. Pharmaceutically active constituents extracted from P. cocos, including polysaccharides, triterpene derivatives, lanostane derivatives, and poricoic acid, exhibited anti-oxidant [12, 13], anti-inflammatory [14], anti-tumor [15–17], anti-emetic [18], anti-nephritic [19], anti-rejection [20], diuretic [21], and anti-hyperglycemic activities [22]. The nematicidal activity of P. cocos was investigated and the active compounds were isolated [23]. Studies on the molecular biology of P. cocos were limited, including the basic molecular studies such as gene expression analysis and gene function identification [24]. qRT-PCR method was effective to detect the candidate genes involved in secondary metabolite biosynthesis. For example, the genes are most likely involved in the biosynthesis of pachymic acid in P. cocos was identified by qRT-PCR [25]; however, contigs and singletons were used instead of reference genes. The stability of potential internal control genes in P. cocos has not been evaluated.
This study aims to discover and obtain the stable reference genes of P. cocos for normalization of qRT-PCR analysis.
Methods
Sampling and culture conditions
The P. cocos strain CGMCC5.78 was purchased from the Institute of Microbiology, Chinese Academy of Sciences and was stored in the Institute of Medicinal Plant Development at −80 °C. We identified the strain using the DNA barcoding method with ITS2 primers. Ninety mycelial samples under different culture conditions were used in this study. Vegetative mycelia were cultured in two different media: potato dextrose agar medium (AOBOX, Beijing, China) and sucrose medium. The components of the sucrose medium were as follows: vitamin B1, 0.05 g/L; MgSO4∙7H2O, 0.5 g/L; KH2PO4∙H2O, 1 g/L; yeast extract, 2.5 g/L; peptone, 5 g/L; and sucrose, 35 g/L. The strain was maintained in potato dextrose agar medium. In the preculture stage, 40-mL sucrose medium was inoculated with mycelia and shaken (Thermo Fisher Scientific 491, Waltham, MA, USA) at 50 rpm in the dark in an incubator at 28 °C for 1 week. Subsequently, all of the mycelia were spread and were shaken at 120 rpm for an additional week in the dark at 28 °C. Finally, all cultures, including the culture broth, were incubated under various conditions (Table 1), including different concentration of hormones and different temperatures for 24 h.
The samples were arbitrarily allocated into six groups for analysis (Table 1). The samples in groups A, B, C, D, and E were cultured in the media supplemented with different concentrations of indole-3-acetic acid (IAA; Sangon, Shanghai, China), abscisic acid (ABA; Sangon), 6-benzylaminopurine (6-BA;Sigma, St Louis, MO, USA), methyl jasmonate (MeJA; Sigma), and gibberellic acid (GA; Sangon), respectively. Group F comprised samples collected from cultures incubated at five different temperatures. The mycelia were collected by double gauze filters (CWBio, Beijing, China). Each experiment was performed in triplicate. A total of 90 samples were collected, and all of the samples were frozen in liquid nitrogen and stored at −80 °C.
Total RNA extraction, DNase treatment, and cDNA synthesis
The liquid nitrogen frozen samples were ground into fine powder by a mortar and pestle. The total RNA of each sample was extracted by the Polysaccharide and Polyphenol Total RNA Isolation Kit (spin column; BioTeke, Beijing, China) according to the manufacturer’s instructions. The total RNA integrity and quality were confirmed by 1 % agarose gel electrophoresis by ethidium bromide staining. The RNA concentration was determined by a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). One microgram of total RNA of each sample was reverse transcribed by the FastQuant RT Kit (with gDNase; TIANGEN, Beijing, China) according to the manufacturer’s protocol. All templates were diluted 30-fold for PCR and qRT-PCR.
Candidate gene selection, primer design, and validation
Based on previous studies [1, 3, 4] of reference genes determined in other species, 14 genes were evaluated in the present study, including multiple-copy genes. These genes include glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mitogen-activated protein kinase (MAPK), beta actin (β-Act), RNA polymerase subunit2 (RPB2), RNA polymerase subunit1 (RPB1), histone 3 (his3), adenine phosphoribosyl transferase (APT), S-adenosyl methionine decarboxylase (SAMDC), ribosomal protein (RP), beta tubulin (β-Tub), eukaryotic translation initiation factor (EIF), and cyclophilin (CYP). The primer sequences, amplicon size and number of gene copies in the genome are summarized in Table 2. The candidate genes were selected from the P. cocos genome sequence database (SRA: PRJNA42921) by the BLASTP program (National Library of Medicine, USA) and a threshold E-value <1 × 10−50.
Primer Premier 6.0 (PREMIER Biosoft, USA) and DNAMAN (LynnonBiosoft, USA) were used for primer design with the following criteria: an amplicon size ranging from 130 to 180 bp, an optimal T m of 53–55 °C, and a primer length from 18 to 22 bp. The primers were synthesized by Sangon Biotech (Shanghai, China). The specificity of each primer pair was measured by 2 % agarose gel electrophoresis following PCR (95 °C for 5 min; 35 cycles of 95 °C for 15 s and 60 °C for 1 min; 72 °C for 10 min) by the 90 cDNA sample mixture. Additionally, qRT-PCR was performed and the melting curve was determined for primers specific validation.
Real-time PCR performance and Ct data collection
The expression level of each gene was determined in 96-well plates by an Applied Biosystems 7500 Real-Time PCR system (Life Technologies, Grand Island, NY, USA). Each reaction mixture contained 200 nM of each primer, 2 µL of the prepared cDNA template, 4.9-µL ddH2O, and 7.5-µL Ultra SYBR Mixture with ROX (CWBio, Beijing, China) in a final volume of 15 µL. The amplifications were performed by an initial denaturation step of 95 °C for 5 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. A temperature ramp step was added after 45 amplification cycles for specificity analysis (melting curve), with 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s, and a final temperature of 60 °C for 15 s. There were three biological duplicate samples, and each biological duplicate sample was evaluated in triplicate.
Data analysis
The Ct values from each reaction were used for analysis of the expression levels of all detected reference genes. The geNorm [26], NormFinder [27], BestKeeper [28], the Delta CT method [29] and the Web-based tool RefFinder [30] were used to determine the stability of the candidate reference genes. The default parameters of these software were applied.
Results
Expression profile of candidate reference genes
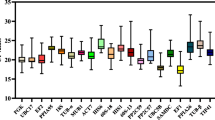
The mean Ct value was computed by three biological duplicates and three technical replicates for each independent experiment (the template generated from each condition of P. cocos was used in different independent experiment), and the three technical replicates were performed independently. A higher Ct value indicates decreased transcription of the target gene. The average Ct value of each candidate gene under conditions ranged from 22.45 ± 0.97 to 32.86 ± 0.86 cycles (Table 3). The average Ct value of six of the 14 genes was higher than 30.00. RPB1-2 and CYP demonstrated the lowest and highest relative expression levels, with average Ct values of 31.21–33.21 and 22.37–23.91, respectively. The variation in the Ct value was determined by the maximum and minimum Ct values. The variation in the Ct value of each candidate reference gene in all 90 samples ranged between 3.22 and 7.89. RPB1-1 exhibited the lowest variation in Ct value followed by CYP (3.24). In contrast, EIF exhibited the highest variation in Ct value (7.89).
Stability ranking of candidate reference genes
geNorm ranks the potential reference genes on the basis of their average pairwise variation in expression of one gene compared with each other gene of the set [26]. geNorm recommends 1.5 as the M-value cutoff. An M-value of less than 1.5 indicates stable expression, with the lowest M-value corresponding to the highest stability, and vice versa. Two reference genes were recommended for an ideal relative quantitative analysis. The M-values of candidate genes under different conditions generated by geNorm are listed in Table 4. The stability of the genes under different treatment conditions analyzed by geNorm is shown in Figs. 1, 2, 3. In group A, his3-1 and CYP were the most stable genes, and his3-2 was the most unstable gene. In group B, MAPK and EIF exhibited the highest stability, and β-Act exhibited the lowest stability. Under different temperatures, the expression levels of RPB1-2 and RPB2 were the most stable in the cultured mycelia, and SAMDC exhibited a performance that was worse than those of the other 13 genes. When treated with different concentrations of 6-BA, RP and CYP were the most stable reference genes, and RPB1-2 exhibited the highest M-value. Following treatment with MeJA, an inducer of secondary metabolism [5, 6], the best reference genes were RPB1-2 and his3-1, whereas GAPDH was the most unstable. Following GA treatment, the expression level of MAPK exhibited the most variation, whereas RPB2 and SAMDC were considered suitable reference genes. Following treatment with IAA, ABA, different temperatures, 6-BA, MeJA, and GA, the average M-values were 0.517–0.871, 0.768–1.36, 0.734–1.242, 0.857–1.333, 0.521–0.849, and 0.602–1.058, respectively. These values suggested that 6-BA contributed the most to the variation in expression levels of the reference genes. A geNorm analysis using all samples indicated that his3-1 and RP were the most stable genes with the lowest M-values, and APT was the most variable gene.
Stability values of the candidate reference genes calculated using geNorm under different treatment conditions. a, b, c, d, and e Stability values of the 14 candidate genes treated with IAA, ABA, 6-BA, MeJA, and GA, respectively. f Stability values of the14 candidate genes under different temperatures
Determination of the optimal number of control genes for normalization under different treatment conditions. If the V 2/V 3 value is less than 0.15, two genes are suitable for normalization as recommended by geNorm. Otherwise, more genes are needed until the V n /V n+1 value is less than 0.15, and the suitable gene number for normalization is n. a, b, c, d, and e Optimal number of control genes for normalization after IAA, ABA, 6-BA, MeJA, and GA treatment, respectively. f Optimal number of control genes for normalization under different temperatures
NormFinder is an Excel-based program for evaluating the expression stability of candidate reference genes based on the expression values, which enables estimation not only of the overall variation of the candidate normalization genes but also of variation between sample subgroups of the sample set [27]. NormFinder shows less sensitivity toward coregulation of the candidate normalization genes. A lower stability value indicates a higher stability. In group A, SAMDC was the most stable gene, with a stability value of 0.135, whereas his3-2 was the most unstable gene, with a stability value of 0.769. In group B, his3-2 exhibited the lowest stability value of 0.088, and β-Act exhibited the highest stability value of 1.586. Under different temperatures, his3-2 was the most stable, with a stability value of 0.069, and SAMDC was the least stable, with a stability value of 1.428. In group C, RP exhibited the best performance with a stability value of 0.183, and the expression level of RPB1-2 varied the most under different concentrations of 6-BA, with a stability value of 1.228. Following MeJA treatment, RPB2 exhibited the lowest variation, with a stability value of 0.106, and GAPDH exhibited the lowest stability value of 0.887. In group E, his3-1 was recommended as the reference gene for GA treatment, with a stability value of 0.244, and MAPK was the most unstable gene among the 14 genes, with a stability value of 1.119. When all of the samples were analyzed, CYP exhibited the lowest stability value of 1.320, whereas EIF exhibited the highest stability value at 13.240.
Gene expression stability was evaluated by BestKeeper using the standard deviation (SD), percentage covariance (CV), and correlation coefficient (r) [28]. BestKeeper can determine the best suited standards, out of 10 candidates, and combine them into an index. The candidate reference genes with SD >1 are considered unstable, and a higher SD value indicates greater variation. From groups A to F, the recommended reference genes were β-Tub, CYP, RP, his3-2, MAPK, and EIF, with SD values of 0.397, 0.171, 0.234, 0.420, 0.345, and 0.297, respectively. In addition, the genes exhibiting the highest SD values in groups A, B, C, D, E, and F were his3-2 (SD = 1.115), RPB1-2 (SD = 1.137), GAPDH (SD = 0.842), MAPK (SD = 0.949), RP (SD = 1.468), and RP (SD = 1.491), respectively. When all 90 samples were considered, the expression level of CYP was the most stable, whereas EIF was the most unstable, with SD values of 0.608 and 1.980, respectively.
RefFinder analysis integrates four different methods (i.e., Delta CT, geNorm, NormFinder, and BestKeeper). The Ct values were input into RefFinder directly, and the ranking of the four methods was calculated. Based on the rankings from each method, RefFinder assigns an appropriate weight to an individual gene and calculates the geometric mean of their weights for the overall final ranking [30]. The rankings of the candidate reference genes used in Delta CT were according to the repeatability of the gene expression differences among the samples. The results analyzed by RefFinder are summarized in Tables 5, 6, 7, 8, 9, 10. In group A, SAMDC was recommended as the most stable reference gene. In group B, his3-2 exhibited the best performance, whereas his3-1 exhibited the best performance in group E. Under different treatment temperatures and different concentrations of MeJA, RPB2 maintained a stable expression level. Following 6-BA treatment, RP ranked as a suitable reference gene (Table 7). Following comprehensive analysis of all of the samples under the various conditions by Delta CT, geNorm, NormFinder, and BestKeeper, CYP was recommended as the reference gene.
The results obtained using these different methods were not identical. In group C, RP was recommended as the most stable gene by all of these methods, whereas in group F, RPB2 was recommended as the reference gene by Delta CT, geNorm and RefFinder. However, in the remaining groups, Delta CT, NormFinder and RefFinder recommended the same gene as the reference gene; SAMDC, his3-2, RPB2 and his3-1 in groups A, B, D, and E, respectively. Following comprehensive analysis of all of the samples under the various conditions, CYP was recommended as the reference gene by Delta CT, BestKeeper, NormFinder, and RefFinder, although not with geNorm. According to the above-mentioned results, RefFinder was likely the most comprehensive and scientific of these methods.
Evaluation of the combination of reference genes
Pairwise variation (V) determines the optimal number of control genes for normalization and proposes 0.15 as a cutoff value [26]. If the V n /V n+1 value is less than 0.15, the suitable gene number for normalization is n. Additional control genes were not necessary in the six groups except for group C (i.e., the 6-BA treatment group),as indicated by V 2/V 3 values below 0.15 [26]. Three reference genes were recommended for group C, as indicated by a V 3/V 4 value of 0.115, which is consistent with the M-value ranking for this group.
Discussion
Validation of the stability of candidate reference genes under different experimental conditions [31], with different tissues [32, 33], at different stages, and in different species [34] is necessary. In the present study, EIF was the most unstable gene in P. cocos; however, EIF1 and EIF3 were recommended as reference genes in Ammopiptanthus mongolicus (Maxim. ex Kom.) S.H. Cheng [35]. In contrast, CYP was the most stable gene in leaves of Deschampsia antarctica É. Desv. [36] under three abiotic stresses (salt, cold, and PEG treatment), whereas the EF-1α gene was recommended for roots. In banana fruit, the expression levels of two widely used reference genes, actin and GAPDH, were not stable [34].
The candidate reference gene rankings for the individual groups evaluated in this study may differ slightly from the ranking for all samples because, under specific circumstances, more accurate rankings would be established. Moreover, most of the M-values of the 14 genes were less than 1.5 except for SAMDC, EIF, and APT, indicating that most of the candidate reference genes were stable. As one of the least stable genes, the instability of APT has been reported in papaya under six experimental conditions [37]. It was contradictory that CYP was the best overall reference gene but did not exhibit the best performance in any single group. CYP was the most stable reference gene using Delta CT, NormFinder, and BestKeeper but not geNorm (Table 11). In addition, CYP was the third-most stable reference gene by geNorm. Moreover, CYP frequently ranked among the top five reference genes (Tables 5, 6, 7, 8, 9, 10), particularly under 6-BA treatment, in which CYP exhibited the highest average M-value when using geNorm for analysis. In group C, CYP ranked firmly as the second-most stable reference gene. In contrast, the ranking of other candidate genes in the six groups varied greatly. A similar phenomenon has been observed in Ammopiptanthus mongolicus [35]. EIF1 and EIF3 were selected as reference genes across all of the samples, whereas these two genes were the most stable only under drought stress among the four evaluated abiotic stresses. Following acibenzolar-S-methyl treatment, the combination of CYP and eIF4B was most suitable as an internal control in Eucalyptus L’Hér. In addition to P. cocos and Eucalyptus [38], CYP has been selected as an internal control for several animal cells. In human peripheral blood, CYP was a more suitable housekeeping gene than β-Act and GAPDH [39]. CYP was also recommended as one of the reference genes for neurons of the central nervous system [40] and in atopic human bronchial epithelial cells [41]. Moreover, CYP was considered to be an RNA normalization control in rats [42].
NormFinder, BestKeeper and geNorm are widely used for selection of reference genes, although the results generated by the different methods may be slightly different [43, 44]. Our results displayed the same tendency as those of previous studies [26–30]. Moreover, the validity of the results might be related to the materials used or even to potential experimental errors. The importance of systematic evaluation before candidate genes are used as reference genes, especially under different conditions were observed in the study.
Conclusion
SAMDC, his3-2, RP, RPB2, and his3-1 were evaluated to be suitable reference genes for P. Cocos following different treatments. RPB2 was the most stable reference gene under different temperatures and CYP was the most stable gene in the mycelia under all six evaluated conditions.
Abbreviations
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- MAPK:
-
mitogen-activated protein kinase
- β-Act:
-
beta actin
- RPB2:
-
RNA polymerase subunit 2
- RPB1-1:
-
RNA polymerase subunit 1
- RPB1-2:
-
RNA polymerase subunit 1
- his3-1:
-
histone 3
- his3-2:
-
histone 3
- APT:
-
adenine phosphoribosyl transferase
- SAMDC:
-
S-adenosyl methionine decarboxylase
- RP:
-
ribosomal protein
- β-Tub:
-
beta tubulin
- EIF:
-
eukaryotic translation initiation factor
- CYP:
-
cyclophilin
- IAA:
-
indole-3-acetic acid
- ABA:
-
abscisic acid
- 6-BA:
-
6-benzylaminopurine
- MeJA:
-
methyl jasmonate
- GA:
-
gibberellin acid
- qRT-PCR:
-
quantitative real-time PCR
- PCR:
-
polymerase chain reaction
References
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17.
Sun Y, Li Y, Luo D, Liao DJ. Pseudogenes as weaknesses of ACTB (Actb) and GAPDH (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PLoS ONE. 2012;7:e41659.
Marum L, Miguel A, Ricardo CP, Miguel C. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE. 2012;7:e35113.
Chooi WH, Zhou R, Yeo SS, Zhang F, Wang DA. Determination and validation of reference gene stability for qPCR analysis in polysaccharide hydrogel-based 3D chondrocytes and mesenchymal stem cell cultural models. Mol Biotechnol. 2013;54:623–33.
Ren A, Qin L, Shi L, Dong X, da Mu S, Li YX, Zhao MW. Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour Technol. 2010;101:6785–90.
Shang CH, Shi L, Ren A, Qin L, Zhao MW. Molecular cloning, characterization, and differential expression of a lanosterol synthase gene from Ganoderma lucidum. Biosci Biotech Bioch. 2010;74:974–8.
Vieira A, Talhinhas P, Loureiro A, Duplessis S, Fernandez D, Silva Mdo C, Paulo OS, Azinheira HG. Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biol. 2011;115:891–901.
Steiger MG, Mach RL, Mach-Aigner AR. An accurate normalization strategy for RT-qPCR in Hypocrea jecorina (Trichoderma reesei). J Biotechnol. 2010;145:30–7.
Xu J, Xu Z, Zhu Y, Luo H, Qian J, Ji A, Hu Y, Sun W, Wang B, Song J, et al. Identification and Evaluation of Reference Genes for qRT-PCR Normalization in Ganoderma lucidum. Curr Microbiol. 2014;68:120–6.
Zhao YY, Feng YL, Bai X, Tan XJ, Lin RC, Mei Q. Ultra performance liquid chromatography-based metabonomic study of therapeutic effect of the surface layer of Poria cocos on adenine-induced chronic kidney disease provides new insight into anti-fibrosis mechanism. PLoS ONE. 2013;8:e59617.
Mantovani MS, Bellini MF, Angeli JP, Oliveira RJ, Silva AF, Ribeiro LR. beta-Glucans in promoting health: prevention against mutation and cancer. Mutat Res. 2008;658:154–61.
Wu SJ, Ng LT, Lin CC. Antioxidant activities of some common ingredients of traditional chinese medicine, Angelica sinensis, Lycium barbarum and Poria cocos. Phytother Res. 2004;18:1008–12.
Zhou L, Zhang Y, Gapter LA, Ling H, Agarwal R, Ng KY. Cytotoxic and anti-oxidant activities of lanostane-type triterpenes isolated from Poria cocos. Chem Pharm Bull. 2008;56:1459–62.
Yasukawa K, Kaminaga T, Kitanaka S, Tai T, Nunoura Y, Natori S, Takido M. 3 beta-p-hydroxybenzoyldehydrotumulosic acid from Poria cocos, and its anti-inflammatory effect. Phytochemistry. 1998;48:1357–60.
Lin Y, Zhang L, Chen L, Jin Y, Zeng F, Jin J, Wan B, Cheung PC. Molecular mass and antitumor activities of sulfated derivatives of alpha-glucan from Poria cocos mycelia. Int J Biol Macromol. 2004;34:289–94.
Ukiya M, Akihisa T, Tokuda H, Hirano M, Oshikubo M, Nobukuni Y, Kimura Y, Tai T, Kondo S, Nishino H. Inhibition of tumor-promoting effects by poricoic acids G and H and other lanostane-type triterpenes and cytotoxic activity of poricoic acids A and G from Poria cocos. J Nat Prod. 2002;65:462–5.
Gapter L, Wang Z, Glinski J, Ng KY. Induction of apoptosis in prostate cancer cells by pachymic acid from Poria cocos. Biochem Biophys Res Commun. 2005;332:1153–61.
Tai T, Akita Y, Kinoshita K, Koyama K, Takahashi K, Watanabe K. Anti-emetic principles of Poria cocos. Planta Med. 1995;61:527–30.
Hattori T, Hayashi K, Nagao T, Furuta K, Ito M, Suzuki Y. Studies on antinephritic effects of plant components (3): effect of pachyman, a main component of Poria cocos Wolf on original-type anti-GBM nephritis in rats and its mechanisms. Jpn J Pharmacol. 1992;59:89–96.
Zhang GW, Liu HY, Xia QM, Li JQ, Lu H, Zhang QH, Yao ZF. Anti-rejection effect of ethanol extract of Poria cocos wolf in rats after cardiac allograft implantation. Chin Med J. 2004;117:932–5.
Zhao YY, Feng YL, Du X, Xi ZH, Cheng XL, Wei F. Diuretic activity of the ethanol and aqueous extracts of the surface layer of Poria cocos in rat. J Ethnopharmacol. 2012;144:775–8.
Li TH, Hou CC, Chang CL, Yang WC. Anti-hyperglycemic properties of crude extract and triterpenes from Poria cocos. Evid-based Complement Alternat Med. 2011;2011.
Li GH, Shen YM, Zhang KQ. Nematicidal activity and chemical component of Poria cocos. J Microbiol. 2005;43:17–20.
Wang JR, Lin JF, Guo LQ, You LF, Zeng XL, Wen JM. Cloning and characterization of squalene synthase gene from Poria cocos and its up-regulation by methyl jasmonate. World J Microb Biot. 2013.
Shu S, Chen B, Zhou M, Zhao X, Xia H, Wang M. De novo sequencing and transcriptome analysis of Wolfiporia cocos to reveal genes related to biosynthesis of triterpenoids. PLoS ONE. 2013;8:e71350.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3: RESEARCH0034.
Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15.
Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33.
Xie F, Sun G, Stiller JW, Zhang B. Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS ONE. 2011;6:e26980.
Toegel S, Huang W, Piana C, Unger FM, Wirth M, Goldring MB, Gabor F, Viernstein H. Selection of reliable reference genes for qPCR studies on chondroprotective action. BMC Mol Biol. 2007;8:13.
Soes S, Sorensen BS, Alsner J, Overgaard J, Hager H, Hansen LL, Kristensen LS. Identification of accurate reference genes for RT-qPCR analysis of formalin-fixed paraffin-embedded tissue from primary non-small cell lung cancers and brain and lymph node metastases. Lung Cancer. 2013;81:180–6.
Nygard AB, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007;8:67.
Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta. 2011;234:377–90.
Shi J, Liu M, Shi J, Zheng G, Wang Y, Wang J, Chen Y, Lu C, Yin W. Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep. 2012;31:1245–54.
Lee H, Kim JH, Park M, Kim I-C, Yim JH, Lee HK. Reference genes validation for qPCR normalization in Deschampsia antarctica during abiotic stresses. Antarct Sci. 2010;22:477–84.
Zhu X, Li X, Chen W, Chen J, Lu W, Chen L, Fu D. Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE. 2012;7:e44405.
Boava LP, Laia ML, Jacob TR, Dabbas KM, Goncalves JF, Ferro JA, Ferro MI, Furtado EL. Selection of endogenous genes for gene expression studies in Eucalyptus under biotic (Puccinia psidii) and abiotic (acibenzolar-S-methyl) stresses using RT-qPCR. BMC Res Notes. 2010;3:43.
Pachot A, Blond JL, Mougin B, Miossec P. Peptidylpropyl isomerase B (PPIB): a suitable reference gene for mRNA quantification in peripheral whole blood. J Biotechnol. 2004;114:121–4.
Chen J, Sochivko D, Beck H, Marechal D, Wiestler OD, Becker AJ. Activity-induced expression of common reference genes in individual cns neurons. Lab Invest. 2001;81:913–6.
He JQ, Sandford AJ, Wang IM, Stepaniants S, Knight DA, Kicic A, Stick SM, Pare PD. Selection of housekeeping genes for real-time PCR in atopic human bronchial epithelial cells. Eur Respir J. 2008;32:755–62.
Weisinger G, Gavish M, Mazurika C, Zinder O. Transcription of actin, cyclophilin and glyceraldehyde phosphate dehydrogenase genes: tissue- and treatment-specificity. Biochim Biophys Acta. 1999;1446:225–32.
Ceelen L, De Spiegelaere W, David M, De Craene J, Vinken M, Vanhaecke T, Rogiers V. Critical selection of reliable reference genes for gene expression study in the HepaRG cell line. Biochem Pharmacol. 2011;81:1255–61.
Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H. Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta. 2013;237:873–89.
Authors’ contributions
JX designed the study. XZ and ZCX performed RNA extraction. XZ performed qRT-PCR. XZ, ZCX, AJJ, CS and YLH analyzed the data. XZ, JYS, HML and SLC wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Dr. Xu Guo (Shandong Academy of Agricultural Sciences) is acknowledged for providing the P. cocos strain CGMCC5.78 used in this work. This work was supported by the National Key Technology R&D Program (Grant no. 2012BAI29B01) and the Program for Innovative Research Team at the Institute of Medicinal Plant Development (IMPLAD).
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, X., Xu, ZC., Xu, J. et al. Selection and validation of reference genes for normalization of quantitative real-time reverse transcription PCR analysis in Poria cocos (Schw.) Wolf (Fuling). Chin Med 11, 8 (2016). https://doi.org/10.1186/s13020-016-0079-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-016-0079-8