Abstract
Background
Recently, a novel group of CD34 and S100 co-expression spindle cell tumors with distinctive stromal and perivascular hyalinization harboring recurrent gene fusions involving RET, RAF1, BRAF, and NTRK1/2 gene has been identified.
Case presentation
In this study, we reported two Chinese male patients with soft tissue tumors presenting in the right knee joint and the left thigh, respectively. For both patients, the tumors were completely excised with clear margin. Microscopically, case 1showed morphological overlap with neurofibroma, and case 2 showed overlap with lipomatous solitary fibrous tumor. Both tumors showed co-expression of S100 and CD34, and absence of SOX10. Genomic profiling with DNA-based next-generation sequencing (NGS) assay was performed and revealed KIF5B-RAF1 (K16:R8) and TLN2-RAF1 (T54:R8) rearrangements. RNA-based NGS and RT-PCR were performed to confirm the gene fusion.
Conclusions
Though systemic therapy was not indicated in these two patients, identification of targetable kinase fusions may help to refine tumors with an ambiguous immunoprofile, and provides suggestions for targeted therapy in rare aggressive cases.
Similar content being viewed by others
Background
Recently, a series of low to intermediate grades of soft tissue tumors showing recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes have been defined as a new entity with S100 and CD34 co-expression (without SOX-10 positivity) (1). These tumors exhibit a spectrum of histological features, including monomorphic spindle cell proliferation, “patternless” growth pattern, stromal and perivascular hyalinization. Herein, we report two morphologically distinct, S100 and CD34 co-expressed low-grade soft tissue tumors carrying novel RAF1 gene fusions with kinesin family member 5B (KIF5B) and talin2 (TLN2), respectively.
Case presentation
Case 1
was a 38-year-old male who presented with an asymptomatic, painless mass in his right knee joint. Ultrasound revealed a 7.7 × 6.2 × 2.5 cm irregular hypoechoic mass, with unclear boundary and abundant blood flow signals inside. Magnetic resonance imaging (MRI) confirmed a lobulated intramuscular mass in his right thigh. Case 2 was a 15-year-old boy who complained of an asymptomatic, painless mass in his left thigh. For both patients, the tumors were completely excised with a clear margin. Subsequent 9-month follow-up of case 1and 6-month follow-up of case 2 did not reveal local or distant recurrence by clinical examination and MRI.
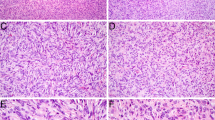
Among soft tissue tumors with CD34 and S100 expression, neurofibroma and malignant peripheral nerve sheath tumor (MPNST) are the main differential diagnosis. In case1 , mucinous degeneration was obvious. Short spindle cells were scattered. The tumor showed the morphological characteristic of neurofibroma (Fig. 1 A). In case 2, the spindle cells proliferated diffusely with no obvious atypia. Mitotic figures were rare. The tumor was composed of mature adipocytes admixed with spindle cells, showing lipomatous solitary fibrous tumor (LSFT) like morphology (Fig. 1 B). Immunohistochemically, both cases shared the same profile. The tumors showed diffuse and strong expression of S100 protein (Fig. 1 C,D), CD34 (Fig. 1E F), and absence of SOX10 positivity. The cells also showed positive staining for vimentin, H3K27Me3, CD31, and SMA. Immunostains were negative for pan-TRK, CK, EMA, ERG, CD68, Desmin, MyoD1, Myogenin, TLE1 and STAT6. Furthermore, Ki67 index of the patient 1 and patient 2 samples were 20% and 10%, respectively. We considered that both patients had a low-grade soft tissue tumor with S100 and CD34 co-expression.
Histologic and immunohistochemical features of RAF1-rearranged tumors. A, Neurofibroma like tumor in a 30-year-old male showing slatternly arranged short spindle cells with striking perivascular rings of collagen (case1 ; ×20). B, LSFT -like tumor in a 15-year-old male showing diffusely arranged spindle cells without obvious atypia and mitotic figures (case 2; ×20). C, E, Immunohistochemical studies showing diffuse S100 and CD34 positivity (case1 ; ×20). D, F, Immunohistochemically, the tumor cells showed diffuse expression of S100 and CD34 (case 2; ×20)
Given the uncertainty in diagnosis, genomic alternations were studied with both DNA-based next-generation sequencing (NGS) assay (Burning Rock OncoScreen Plus ®, Burning Rock Biotech, Guangzhou, China) and targeted RNA platform (OncoRNA, Burning Rock Biotech, Guangzhou, China) to identify additional molecular alterations and to potentially aid in patient management. DNA- and RNA-based NGS identified gene fusions of RAF1 exon 8 to KIF5B exon 16 in case1 , and RAF1 exon 8 to TLN2 exon 54 in case 2. In case1 , DNA-based NGS identified a KIF5B-RAF1 (K16:R8) rearrangement resulting from an in frame t(10;3) (q21.3; q11.2) translocation. RNA-based NGS confirmed these uncommon RAF1 rearrangements from DNA sequencing, and additionally identified a reciprocal RAF1- KIF5B (R7:K17) fusion transcript (Fig. 2 A-C). The KIF5B-RAF1 and RAF1- KIF5B fusions were further confirmed by reverse-transcriptase polymerase chain reaction (RT-PCR; Fig. 2D). In case 2, DNA-based NGS identified a TLN2-RAF1 (T54:R8) rearrangement resulting from an in-frame t(15;3) (q21.1; q11.2) translocation, which was confirmed by RNA-based NGS (Fig. 3 A, B). Based on the clinicopathologic and genomic features of the two cases, RAF 1 fusion-positive soft tissue tumor with S100 and CD34 co-expression was the final diagnosis.
Genomic structures of the KIF5B-RAF1 and RAF1-KIF5B fusions identified in tumor are shown based on RNA-based NGS. A, B, Breakpoints were detected in intron 8 of KIF5B and intron 3 of RAF1, separately. The predicated resultant gene fusions were between exon 16 of KIF5B and exon 8 of RAF1, and exon 7 of RAF-1 and exon 17 of KIF5B. C. Schematic representation of the KIF5B-RAF1 and RAF1-KIF5B fusion transcripts. D, RT-PCR amplicon product revealed the KIF5B -RAF1 and RAF1-KIF5B fusion transcripts were at right size (170 bp, 221 bp; red arrows)
Genomic structure of the TLN2-RAF1 fusion identified in tumor is shown based on RNA-based NGS. A, Breakpoints were detected in intron 54 of TLN2 and intron 7 of RAF1, separately. The predicated resultant gene fusion was between exon 54 of TLN2 and exon 8 of RAF1. C. Schematic representation of the TLN2-RAF1 fusion transcript
Discussion and conclusions
RAF1 is a member of the RAF family of signaling kinases, downstream of RAS, which activates the MEK/ERK pathway that promotes cell proliferation and survival. In soft tissue tumors with S100 and CD34 co-expression, at least 4 RAF1 fusion partner genes have been identified to date, including PDZRN3, SLMAP, and TMF1 genes located on chromosome 3 and MTAP gene located on chromosome 9 (1–3). TLN2 on chromosome 15 and KIF5B on chromosome 10 are first identified as RAF1 fusion partners in our study. Both KIF5B-RAF1 and TLN2-RAF1 fusion transcripts conserve the RAF1 tyrosine kinase domain with the loss of the autoinhibitory region upstream. KIF5B and TLN2 genes retained their promoters to initiate transcription, which then leads to the activation of RAF1 and the downstream MAPK/ERK pathway. KIF5B has been reported as a partner gene in lung adenocarcinomas (LADCs), typically in relation to pericentric inversion events of the RET proto-oncogene (4). TLN2 has been rarely reported as a partner gene in tumors.
Our cases revealed the genetic diversity of RAF1 fusions, and also underscored the need for diagnostic strategies that are fusion partner non-specific. Common methods for detecting gene fusions in the clinic include RT-PCR, break-apart fluorescence in situ hybridization (FISH), and DNA/RNA-based NGS assay. The main advantage of RT-PCR lies in the possibility of identifying specific known RAF1 fusion partners. The performance of RT-PCR is limited by atypical RAF1 rearrangement with new fusion partners and RNA quality. Break-apart FISH plays a critical role in fusion screening tests given its single-cell resolution and rapid turn-around time. Positive FISH results with unusual, complex rearranged patterns and borderline-negative FISH results may benefit from further confirmation with RNA-sequencing. DNA-based NGS is preferable to FISH as a screen due to higher sensitivity for all gene fusions including exons and introns near the kinase domain. However, any atypical RAF1 rearrangements with new fusion partners should be further evaluated by RNA sequencing to confirm the presence of the fusion transcript. The utility of RNA sequencing for confirming unusual rearrangements from DNA sequencing or break-apart FISH could improve the quality and accuracy of RAF1 fusion testing.
In conclusion, we reported two cases of RAF1 fusion transcripts in soft tissue tumor with CD34 and S100 co-expression, extending the molecular genetic spectrum of this recently described entity. Though systemic therapy was not indicated in these two patients, identifying targetable kinase fusions involving the MAPK/ERK pathway may help refine tumors with an ambiguous immunoprofile, and provide new paths for precision medicine strategies involving targeted kinase inhibitors.
Data Availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- NGS:
-
next-generation sequencing.
- MRI:
-
Magnetic resonance imaging.
References
1. Suurmeijer AJH, Dickson BC, Swanson D, Zhang L, Sung YS, Cotzia P, et al. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Genes Chromosomes Cancer. 2018;57(12):611 − 21.
2. Hicks JK, Henderson-Jackson E, Duggan J, Joyce DM, Brohl AS. Identification of a novel MTAP-RAF1 fusion in a soft tissue sarcoma. Diagnostic Pathology. 2018;13(1):77.
3. Mok Y, Kimpo MS, Chen H, Kuick CH, Chang KT, Lee VKM. Spindle cell tumour with S100 and CD34 co-expression showing PDZRN3-RAF1 rearrangement - a recently described entity. Histopathology. 2019;74(7):1109-11.
4. Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18(3):375-7.
Acknowledgements
The authors thank Dr. Xi Li (Burning Rock Biotech, Guangzhou, China), and Dr. Yang Wang (Burning Rock Biotech, Guangzhou, China) for discussing the results.
Funding
None.
Author information
Authors and Affiliations
Contributions
G-LH carried out the data statistic and wrote the manuscript. G-LH, L-WF and N-XH organized the clinical and radiographic materials, and performed immunohistochemical staining and RT-PCR experiments. G-LH and D-Y made the diagnosis. D-Y offered financial support. D-Y revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Written informed consent was obtained from the patient for publication. All authors are in agreement for the publication of the study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gong, LH., Liu, WF., Niu, XH. et al. Two cases of spindle cell tumors with S100 and CD34 co-expression showing novel RAF1 fusions. Diagn Pathol 17, 80 (2022). https://doi.org/10.1186/s13000-022-01263-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-022-01263-y