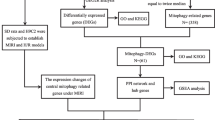
Abstract
Background
Ischemia/reperfusion (I/R) injury of heart is one of the major causes of acute cardiac injury, which may result in worsening or even loss of heart function. With novel microRNAs being evolutionarily discovered, numbers of them remained functionally unknown. We aimed to discover novel microRNAs with therapeutic or diagnostic potential in the setting of early cardiac I/R injury.
Methods
Cardiac electrical activity, biochemical detection and histopathology analysis were performed to reveal early changes of cardiac I/R injury. A microRNA array was performed to screen differential microRNAs in the mouse model of cardiac I/R injury. The differentially expressed microRNAs were validated in cardiac tissues and in serum samples.
Results
The abnormality in electrocardiogram and increases in serum cTnI levels suggested the successful establishment of cardiac I/R injury in mice. A total of 1882 microRNAs were identified, of which 11 were significantly down-regulated and 41 were significantly up-regulated at 3 h post reperfusion. microRNA 223-3p and microRNA 3113-5p were among the mostly altered microRNAs and were validated to be up-regulated within the early hours of I/R injury in heart tissues. In the circulating system, cTnI, a sensitive marker of cardiac injury, or microRNA 223-3p only increased within the first 6 h post I/R injury. However, microRNA 3113-5p stably increased in the serum, keeping an increase of 2.5-fold throughout the 24 h. In the human serum samples, microRNA 3113-5p was detected to be significantly upregulated as soon as 3 h after I/R stimuli and kept significantly higher levels within the 48 h.
Conclusion
This is the first study that reported the functional roles of microRNA 3113-5p in cardiovascular system. Our data suggested that cardiac microRNA 3113-5p might be a useful target for therapeutic purposes and circulating microRNA 3113-5p might serve as a stable marker for early diagnosis of cardiac I/R injury.
Similar content being viewed by others
Background
Ischemia/reperfusion injury (I/R injury) is defined as initial lack of oxygen due to deprivation of blood flow and the following restoration of blood supply from the occluded vessel. I/R injury is a critical mechanism of organ injury [1], and a central cause of tissue injury during various medical conditions, including surgical procedures, organ transplantation, cardiovascular diseases (i.e. myocardial infarction, and circulatory shock) and toxic insults [2]. During the development of I/R injury, a surge of reactive oxygen and nitrogen species as well as inflammatory cascades bursts and eventually triggers organ damage [3]. Other processes such as protein post-translational modifications, lipid oxidations, and DNA breakage are also common mechanisms that aggravate a chain of deleterious responses which eventually result in dysfunction of endothelial cells, neutrophils transmigration to the insulted endothelium, burst of inflammatory cytokines, calcium iron overload, and eventual cell death [4]. Long-term insult by I/R injury could lead to irreversible injury to heart and many studies have demonstrated various therapeutic strategies against long-term I/R injury [5]; however, early diagnosis and treatment of I/R injury, which receives scanty attention, is advantageous over late-stage intervention and thus mandates more research focus.
Recently, microRNAs have been shown to implicate in the pathophysiology of cardiac I/R injury [1]. Multiple microRNAs have been reported to have diagnostic values or therapeutic potentials [6]. For example, microRNA 21 contributed to cardiac diseases by triggering mitogen-activated protein kinase (MAPK) activity in fibroblasts [7]. MicroRNA 21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation [8]. microRNA 223-3p is another widely documented microRNA which has been reported to regulate cardiac fibrosis after myocardial infarction by targeting RAS p21 protein activator 1 (RASA1) [9], and regulated expression of voltage-gated K+ channel Kv4.2 in acute myocardial infarction [10].
Due to the nature of external secretion and functional importance in heart diseases, multiple microRNAs have been anticipated as non-invasive biomarkers. So far, more than 200 microRNAs have been considered as heart-specific with many of them being identified to release into the circulatory system during ischemic injury. It is reported that the combination of microRNA 199a-3p, microRNA 208a-3p, microRNA 122-5p, and microRNA 223-3p has a good diagnostic performance for hypertension [11]. Elevated plasma microRNA 223 content associated with the severity of coronary heart disease [12]. Circulating microRNA 1, 499-5p, and microRNA 133a or the cardiomyocyte-specific microRNA 208b have been reported to instantly increase in patients with the onset of ST-elevated myocardial infarction (STEMI) and their levels peak within 12 h after disease onset [13]. However, despite the numerous reports documenting the potential of microRNAs serving as biomarkers for I/R injury, only limited number of microRNAs have been applied in clinical trials, implicating there is still space to identify novel microRNAs.
One fact is that microRNAs keep expanding and undergo evolutionarily identification [14]. With techniques deeply sequencing microRNAs, some new microRNAs with functionality unknown have been identified [15, 16]. By using the newly refreshed database, an updated microRNA array could provide a panel of microRNAs, some of which might represent new ones that may otherwise remain functionally unknown [15, 16]. The present study aimed to perform an updated microRNA array analysis of heart tissues undergoing I/R injury in order to discover novel microRNAs. The identified microRNAs were then validated through quantitative analysis. Our data provided novel biomarkers for diagnosis and treatment of early cardiac I/R injury.
Methods
Experimental protocol
Male C57BL/J mice (~ 25 g) were initially anesthetized with 5% chloral hydrate and injected with heparin intravenously at a final concentration of 500 U/kg. Mouse heart was then exposed and perfused according to Langendorff with oxygenated, normothermic Krebs-Henseleit buffer as previously described [17]. For the purpose of this study, different reperfusion time was applied to the indicated group of mice. A time-matched nonischemic control group was aerobically reperfused for 190 min. For the details of inducing I/R injury, the left anterior descending coronary artery (LAD) was undergoing continuous occlusion for 45 min to induce regional ischemia. Thereafter, the occluded LAD was reopened to let perfusion with different hours (1 h, 3 h, 6 h, 12 h, or 24 h). Heart rate and coronary flow were real-time monitored during the perfusion periods in all groups. To confirm successful ligation of the LAD, a Data Analysis System (BL-420; TME Technology, Chengdu, Sichuan, China) was used to record the electrocardiogram (ECG) during the I/R period. Samples from all groups (n = 6/group) were immediately frozen in liquid nitrogen for subsequent microRNA isolation. To further assess tissue injury, heart release of cardiac Troponin I (cTnI) was measured using a cTnI ELISA kit (Life Diagnostics Inc., West Chester, PA, USA) from serum samples. In brief, to collect mice serum samples, mice were anesthetized and their hearts were exposed. Following exposure of hearts, the LAD was directly ligated and then reopened for different hours (1 h, 3 h, 6 h, 12 h, or 24 h). In a second series of experiments, the LAD-supplied risk regions of the left ventricle were immediately sampled at the end of 3-h reperfusion and subject to further analysis of differentially expressed microRNAs. For sampling, a single oblique cut was made from the origin of the LAD toward the right side of the apical area. In this way, the samples involve the majority of the left ventricle anterior wall as well as the apex of the heart.
Histopathology analysis
The left ventricle from the LAD-supplied zone was fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin-embedded tissues were then cut into 4 μm-thick slices, which were then stained with hematoxylin & eosin (HE) for histological examination. Briefly, after deparaffinization and rehydration, myocardial sections were sequentially stained with eosin for cytoplasm staining and hematoxylin for nucleus staining. Digital images were obtained at × 200 magnification by microscopy (Olympus, Tokyo, Japan).
Isolation of microRNAs from serum and cardiac tissues
Circulating miRNAs were isolated with the miRNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNAs from heart tissues were isolated using TRIzol® (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA purity and concentration were assessed by spectophotometry at 260 and 280 nm (NanoDrop Products, Wilmington, DE, USA) equipped with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Microarray analysis of microRNA expression
For the purpose of microarray analysis, all microRNAs from heart samples were initially labeled with the microRNA Complete Labeling and Hyb kit system (Agilent Technologies). Labeled samples were then completely vacuum dried at a medium-high temperature (45 °C) and hybridized onto the surface of a mouse microRNA Microarray (Agilent Technologies) in a microarray hybridization chamber (Agilent Technologies) according to previous regulations [18]. The scanning of each array was in accordance with a previous published protocol [19]. Statistical analysis was performed and explicitly described below. Briefly, the fluorescent signal intensity data represented microRNA expression and changes in gene expression were determined as ratios of signal intensity values. For visual comparison and representation of both down- and upregulation, data were processed and depicted as log2 changes.
Analysis of differentially expressed microRNAs were done using Feature Extraction software (Agilent Technologies) as previously described [20]. All individual microRNAs were represented by 20 different probes on the array. A microRNA was considered to be detectable when at least 1 probe from all the 20 probes was detected. The final gene signal equals to the sum of all signals of each individual probe. Using a two-tailed two-sample unequal variance Student’s t-test, the P value was used as a determinate to find significantly expressed microRNAs. A corrected P value was calculated for each microRNA to control the false discovery rate (FDR) using the Benjamini and Hochberg multiple testing correction protocol. MicroRNA expression ratios with P values less than 0.05 and log2 changes of less than − 1.0 or log2 changes of more than 1.0 were considered as significant repression or overexpression, respectively.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Reverse transcription of the total RNA into cDNA was performed using the High Capacity RNA-to-cDNA transcription kit (Applied Biosystems), according to the manufacturer’s instructions. The RT-qPCR assays were performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) following with the TaqMan MicroRNA Assays (Applied Biosystems) according to the manufacturer’s instructions on a 7500 Real-Time RT-qPCR system (Applied Biosystems). The RT-qPCR conditions were as follows: 95 °C for 10 min, and 40 cycles of 95 °C for 13 s and 60 °C for 60 s. The relative expression levels of serum microRNAs were normalized to the relative expression level of U6. The relative expression of microRNAs was calculated using the 2-ΔΔCt equation [21]. Primers used in this study were listed in Table 1. The U6 primer sequences and universal reverse primers were provided by the kits.
We also analyzed the relative expression of predicted target genes of microRNAs. Gapdh was used as the internal control gene for normalization. The primers used were listed in Table 2.
Statistical analysis
Microarray and RT-qPCR data were presented as means ± standard error of mean (SEM). The Students’ t-test was used to compare differences of means between groups. The one-way analysis of variance (ANOVA) was used to evaluate differences among ≥3 groups, which was followed by least-significant-difference (LSD) post hoc test for comparisons within groups. P < 0.05 was considered to be statistically significant.
Results
Early cardiac I/R injury lacks biochemical biomarkers and presented nonspecific histologic changes
Initially, we occluded the left coronary for 45 min and then perfused the artery for different hours (1 h, 3 h, 6 h, 12 h and 24 h) to establish a murine model of cardiac I/R injury. ECG monitoring showed that the mice with LAD occlusion (ischemic mice) exerted remarkable elevation of ST segment, while the ST segment in the sham group of mice remained at the baseline. After 45 min of occlusion, the LAD was re-opened and ECG monitoring showed that the ST segment declined to baseline after 3 h’ perfusion (Fig. 1a), conforming to the manifestation of I/R injury. Moreover, the serum levels of cTnI, a sensitive cardiac injury marker, dramatically increased in the first 3 h after reperfusion; thereafter it dropped but remained significantly higher in the I/R injured mice than that in Sham mice (Fig. 1b). HE staining is the gold standard for diagnosing most of heart diseases. However, despite the significant elevation of cTnI levels during the initial 3 h’ perfusion, histological analysis of heart tissues showed that myocardial cells in the sham group were well-ordered with regular structure. Myocardial cells from the I/R injured mice were swelling in the plasma (black arrow) and showed slight interstitial edema (blue arrow) or occasional cell hypertrophy (green arrow) after 3- or 6-h’ reperfusion. The non-specific histopathological manifestations suggested that histological analysis did not aid in making a diagnosis of cardiac I/R injury.
Establishment of a murine model bearing cardiac ischemia/reperfusion (I/R) injury. a Electrocardiogram monitoring of mice with (ischemia group) or without (Sham group) coronary artery occlusion. Mice after re-opening of the left anterior descending (LAD) artery (perfusion group) were also subject to ECG monitoring. b ELISA detection of serum cTnI levels in Sham group of mice and in mice with distinct perfusion time. c Hematoxylin & Eosin (HE) staining of heart tissues were performed. Myocardial cells in the sham group were well-ordered with regular structure, while myocardial cells from the I/R injured mice were swelling in the plasma (black arrow) and showed slight interstitial edema (blue arrow) or occasional cell hypertrophy (green arrow) after 3 or 6 h’ reperfusion. *, p < 0.05 vs. Sham
microRNA array identified multiple novel microRNAs
To identify sensitive cardiac markers indicating I/R injury, we then performed a microRNA array analysis of hearts tissues with 3 h I/R injury. A total of 1882 microRNAs were identified, of which 11 were significantly down-regulated and 41 were significantly up-regulated at 3 h post reperfusion (Fig. 2a). Scatter plot analysis and volcano plot showed the distribution of all the 1882 microRNAs (Fig. 2b-c). The fold change of repression ranged from 1.6 to nearly 50, while that of upregulation varied from 1.8 to 463 (Fig. 2c). Of great interest, many of the identified microRNAs, such as microRNA 3113-5p, have been only recently discovered and remained largely unknown in function.
Bioinformatics analysis of the differentially expressed microRNAs
Of all these differentially expressed microRNAs, target genes were predicted using four databases including TargetScan, miRDB, miRTarbase and Tarbase. Thereafter, Gene Ontology (GO) analysis revealed that biological processes included multiple processes such as regulation of myosin light chain phosphatase activity, histone H4-K20 methylation and cellular response to nitric oxide. Molecular function included connexin binding, calcium-dependent protein serine/threonine phosphatase activity and voltage-gated sodium channel activity involved in cardiac muscle cells etc. (Fig. 3a). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the majority of target genes were involved in mechanistic target of rapamycin (mTOR) signaling pathway, Axon guidance, and Wnt signaling pathway etc. (Fig. 3b). Since microRNAs function mainly through targeting mRNAs [22, 23], it was hypothesized that microRNAs without valid target genes might not have biological importance. We thus refined microRNAs with target genes predicted by at least 3 databases and concentrated on microRNA 3113-5p, microRNA 5121, microRNA 327, microRNA 1892, microRNA 709, microRNA 1895, microRNA 188-5p, microRNA 1224-5p, microRNA 3968, microRNA 30b-5p, microRNA 202-3p, microRNA 154-5p, microRNA 150-5p, and microRNA 146a-5p (Fig. 3c). Target genes from 3 of the 4 databases were schematically illustrated for all the microRNAs of interest (Fig. 3c).
RT-qPCR validation of interest microRNAs in mice
Based on the above analysis, RT-qPCR analysis of cardiac microRNA expression was then performed. It was initially shown that microRNA 223-3p in the I/R group of mice showed the highest change by up to 4 folds as compared with Sham group of mice, while microRNA 3113-5p was only second to microRNA 223-3p that showed as high as 3-fold upregulation (Fig. 4a). Hence, we further analyzed these two microRNAs in heart tissues with distinct perfusion time. It was found that microRNA 223-3p gradually increased right after perfusion and peaked by 6 h after perfusion. It then remained relatively stable till 24 h after perfusion (Fig. 4b). Moreover, RT-qPCR validation assay showed that microRNA 3113-5p showed stable upregulation during the whole perfusion periods as compared with Sham mice hearts (Fig. 4c). Analysis of the predicted target gene of microRNA 3113-5p, zinc finger protein 933 (Zfp933), showed that it was consistently repressed by 2-folds in the perfused heart tissues, independent of perfusion time (Fig. 4d).
RT-qPCR validation of the differentially expressed microRNAs. a RT-qPCR analysis of all the 14 potential microRNAs in heart tissues. b-c RT-qPCR analysis of microRNA 223-3p and microRNA 3113-5p in heart tissues with distinct perfusion time. d RT-qPCR analysis of cardiac Zfp933 expression (a predicted microRNA 3113-5p target gene) in heart tissues with distinct perfusion time. *, p < 0.05; **, p < 0.01 vs. Sham
Circulating microRNA 3113-5p stably increased in early perfusion hours in mice
In addition to the cardiac expression, we also detected the circulating levels of interest microRNAs. RT-qPCR detection showed that microRNA 223-3p had maximal upregulation in serum samples of perfused mice but declined dramatically after 3 h (Fig. 5a). Unlike microRNA 223-3p, serum levels of microRNA 3113-5p remained approximately 3-fold increased throughout the whole early perfusion time (Fig. 5b).
Circulating microRNA 3113-5p significantly increased in early I/R stimuli in human samples
To further detect the expression of microRNA 3113-5p in human samples, we collected serum samples from a healthy control and 3 patients with cardiac I/R injury (Table 3). All these patients were diagnosed with myocardial injury (MI) and underwent stent implantation. One hour before operation, all patients showed dramatically high levels of cardiac Troponin I (cTnI) and Myoglobin (MYO) and 1 h after operation, they still presented with higher cTnI levels than the normal upper limit (Table 3). The cardiac I/R injury patients were collected sera during time periods of 3 h, 6 h, 24 h, 48 h and 72 h. Our results showed that as soon as 3 h after cardiac I/R stimuli, the circulating microRNA 3113-5p significantly increased by up to 1.5-fold (Fig. 6). The circulating microRNA 3113-5p peaked at 6 h (6-fold) and remained significantly higher than control levels until 48 h after cardiac I/R stimuli. At 72 h after reperfusion, the serum level of microRNA 3113-5p dropped to the basal level as observed in the health control. During the early I/R injury hours (within 48 h), circulating microRNA 3113-5p levels were significantly higher than the healthy control (Fig. 6).
Circulating microRNA 3113-5p significantly increased in the early perfusion hours in human serum samples. Serum samples from healthy people who underwent routine health checkup served as control (Ctrl). Three patients with myocardial infraction and underwent stent implantation were collected sera during time periods of 3 h,6 h, 24 h, 48 h and 72 h. *, P < 0.05, ***, P < 0.0001 vs. Ctrl
Discussion
Organ damage insulted by I/R injury represents a serious medical condition, which often leads to deterioration or even loss of organ function [4]. I/R injury is occurred when transient tissue oxygen deprivation due to vessel occlusion accompanies with a subsequent reperfusion period following recovery of blood supply. During the reperfusion periods, multiple reactive oxygen and nitrogen species accumulate and result in eventual tissue injury [3].
In order to facilitate early diagnosis of acute I/R injury insults and risk stratification for future adverse cardiac events, it is constantly urgent to find cardiac or circulatory biomarkers that refine the diagnosis and management of patients with symptoms related to acute or chronic cardiovascular diseases [1]. microRNAs are a class of noncoding RNAs that negatively affect gene transcription through binding to the mRNA [24]. Multiple microRNAs have been identified to have diagnostic values or therapeutic potentials, but few have been applied into clinical trials. Since noncoding RNAs have been revolving to forge new ones [16], it is necessary to re-perform a microRNA array in order to discover a panel of novel microRNAs.
Since histopathological analysis revealed non-specific and non-significant changes, identification of molecular markers would be of great importance. Using an updated database with refreshed microRNAs, the current study identified 52 microRNAs that differentially expressed after 3 h of I/R injury. GO and KEGG bioinformatics analyses revealed that these microRNAs were associated with regulation of myosin light chain phosphatase activity, histone H4-K20 methylation and cellular response to nitric oxide. Many of these microRNAs were consistent with previous reports such as microRNA 146a [25] and some were inconsistent which might be explained by different I/R injured hours (we used 3 h of I/R injury for microRNA array). Of all these microRNAs, microRNA 223-3p and microRNA 3113-5p showed the highest fold changes. MicroRNA 223 expression was greatly upregulated in the livers after 75 min ischemia followed by 120 min reperfusion and its expression associated with hepatic I/R injury [26]. In a more recent study, microRNA 223-3p and -5p cooperatively suppressed necroptosis in I/R injured hearts [27]. MicroRNA 223-3p also regulated cardiac fibrosis after myocardial infarction by targeting RASA1 [9], and regulated expression of voltage-gated K+ channel Kv4.2 in acute myocardial infarction [10]. Similar with previous reports, the microRNA array also identified that microRNA-223-3p was among the top ones that altered expression, which was in conformation with previous reports that associated microRNA 223-3p with heart diseases [9, 10]. In the validation assay, cardiac microRNA 223-3p kept increasing after reperfusion but its expression in serum samples only peaked at 1 h of perfusion, indicating that microRNA 223-3p might be a valuable biomarker of cardiac I/R injury but this value was only within a very narrow time window.
Though not altered as much as microRNA 223-3p, microRNA 3113-5p was only second to it as evidenced by RT-qPCR validation. Different from microRNA 223-3p, microRNA 3113-5p was stably upregulated in both cardiac tissues and the serum samples. Right after perfusion, the cardiac microRNA 3113-5p level was approximately 2–3 folds increased and kept at this high level throughout the whole perfusion periods. Its predicted target gene, Zfp933, was also stably repressed and kept at a minimum level in distinct perfusion hours. Zfp933 is one member from the zinc finger protein family with its function mysterious. The C2H2-zinc finger proteins constitute the largest class of transcription factors within the human genome. These proteins are generally involved in crucial cell functions, such as survival and growth [28]. It has been reported that Zfp580, a Zfp933 homogenous protein, shows the highest expression in the heart and serves as a cardioprotection factor against cardiac I/R injury [29]. Hence, the identification of Zfp933 as the target gene of microRNA 3113-5p further implicated the functional significance of microRNA 3113-5p and its expression pattern in turn reinforced the stable cardiac increase of microRNA 3113-5p. One great advantage that makes microRNAs superior to other biomarkers is that microRNAs could stably exist in the extracellular space, such as circulating blood, urine, vitreous humor and other body fluids in spite of the existence of RNases [30]. Multiple extracellular microRNAs have been identified to have potential function and pivotal roles as disease biomarkers [31]. We therefore detected serum levels of microRNA 3113-5p in mice and in human samples in order to find non-invasive biomarkers. It was found that the serum levels of microRNA 3113-5p also stably increased ever since perfusion. The increase of microRNA 3113-5p was approximately 2–3 folds in mice serum, a change similar to that in cardiac levels, indicating that the cardiac and circulating levels of microRNA-3113-5p were highly consistent. In human serum samples, microRNA 3113-5p was significantly increased after 3 h of I/R injury and peaked by 6 h and this significant increase could be detected till 48 h. Validation of microRNA 3113-5p expression in human sera further suggested the diagnostic potential in cardiac I/R injury. Therefore, the early increase of microRNA 3113-5p made it a superior biomarker since it might serve as an early and stable indicator for cardiac I/R injury.
MicroRNA 3113-5p is a new microRNA that remains mysterious as to its function. This study represents the first one to report its functional association with cardiac I/R injury. The stable increase in cardiac and circulating microRNA 3113-5p levels suggested that it is a valuable microRNA that may aid in diagnosing I/R injury or serve as a potential therapeutic target in the heart for medical intervention of I/R injury.
Conclusions
The present study re-analyzed the differentially expressed microRNAs during cardiac I/R injury, with an updated microRNA array. Multiple novel microRNAs that remains functionally unknown were identified. Among them, microRNA 223-3p and microRNA 3113-5p were the top ones that were validated to have functional association with I/R injury. microRNA 3113-5p is a stable biomarker that altered significantly in early hours of reperfusion. This is the first study that identified a novel microRNA, microRNA 3113-5p as a critical biomarker for cardiac I/R injury. Cardiac microRNA 3113-5p might be a useful target for therapeutic purposes and circulating microRNA 3113-5p might serve as a stable marker for diagnosis of cardiac I/R injury.
Limitations
The present study only assessed the association of microRNA 3113-5p with cardiac I/R injury. The expression of microRNA 3113-5p in I/R injury from other organs was not detected.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- cTnI:
-
cardiac Troponin I
- ECG:
-
Electrocardiogram
- Gapdh:
-
Glyceraldehyde phosphate dehydrogenase
- GO:
-
Gene ontology
- HE:
-
Hematoxylin & eosin
- I/R injury:
-
Ischemia/reperfusion injury
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LAD:
-
Descending coronary artery
- MAPK:
-
Mitogen-activated protein kinase
- MI:
-
Myocardial infarction
- mTOR:
-
mechanistic target of rapamycin
- MYO:
-
Myoglobin
- NA:
-
Not applicable
- RASA1:
-
RAS p21 protein activator 1
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- STEMI:
-
ST-elevated myocardial infarction
- Zfp933:
-
Zinc finger protein 933
References
Lorenzen JM, Batkai S, Thum T. Regulation of cardiac and renal ischemia-reperfusion injury by microRNAs. Free Radic Biol Med. 2013;64:78–84.
Woodall MC, Woodall BP, Gao E, Yuan A, Koch WJ. Cardiac fibroblast GRK2 deletion enhances contractility and remodeling following ischemia/reperfusion injury. Circ Res. 2016;119(10):1116–27.
Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87(1):315–424.
Hausenloy DJ, Yellon DM. Targeting myocardial reperfusion injury--the search continues. N Engl J Med. 2015;373(11):1073–5.
Wang X, Wang J, Tu T, Iyan Z, Mungun D, Yang Z, Guo Y. Remote ischemic Postconditioning protects against myocardial ischemia-reperfusion injury by inhibition of the RAGE-HMGB1 pathway. Biomed Res Int. 2018;2018:4565630.
Min H, Fan S, Song S, Zhuang Y, Li H, Wu Y, Cai H, Yi L, Dai J, Gao Q. Plasma microRNAs are associated with acute exacerbation in idiopathic pulmonary fibrosis. Diagn Pathol. 2016;11(1):135.
Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S. And others. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–4.
Li H, Zhang X, Wang F, Zhou L, Yin Z, Fan J, Nie X, Wang P, Fu XD, Chen C. And others. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by Upregulating mitochondrial translation. Circulation. 2016;134(10):734–51.
Liu X, Xu Y, Deng Y, Li H. MicroRNA-223 regulates cardiac fibrosis after myocardial infarction by targeting RASA1. Cell Physiol Biochem. 2018;46(4):1439–54.
Liu X, Zhang Y, Du W, Liang H, He H, Zhang L, Pan Z, Li X, Xu C, Zhou Y. And others. MiR-223-3p as a novel MicroRNA regulator of expression of voltage-gated K+ channel Kv4.2 in acute myocardial infarction. Cell Physiol Biochem. 2016;39(1):102–14.
Zhang X, Wang X, Wu J, Peng J, Deng X, Shen Y, Yang C, Yuan J, Zou YZ. The diagnostic values of circulating microRNAs for hypertension and bioinformatic analysis. Biosci Rep. 2018;38:4.
Guo JF, Zhang Y, Zheng QX, Zhang Y, Zhou HH, Cui LM. Association between elevated plasma microRNA-223 content and severity of coronary heart disease. Scand J Clin Lab Invest. 2018;78(5):1–6.
Gholamin S, Pasdar A, Khorrami MS, Mirzaei H, Mirzaei HR, Salehi R, Ferns GA, Ghayour-Mobarhan M, Avan A. The potential for circulating microRNAs in the diagnosis of myocardial infarction: a novel approach to disease diagnosis and treatment. Curr Pharm Des. 2016;22(3):397–403.
Ferracin M, Negrini M. Micromarkers 2.0: an update on the role of microRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2015;15(10):1369–81.
Islas JF, Moreno-Cuevas JE. A MicroRNA Perspective on Cardiovascular Development and Diseases: An Update. Int J Mol Sci. 2018;19(7).
Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94.
Csonka C, Szilvassy Z, Fulop F, Pali T, Blasig IE, Tosaki A, Schulz R, Ferdinandy P. Classic preconditioning decreases the harmful accumulation of nitric oxide during ischemia and reperfusion in rat hearts. Circulation. 1999;100(22):2260–6.
Farago N, Kocsis GF, Feher LZ, Csont T, Hackler LJ, Varga-Orvos Z, Csonka C, Kelemen JZ, Ferdinandy P, Puskas LG. Gene and protein expression changes in response to normoxic perfusion in mouse hearts. J Pharmacol Toxicol Methods. 2008;57(2):145–54.
Yang F, You X, Xu T, Liu Y, Ren Y, Liu S, Wu F, Xu Z, Zou L, Wang G. Screening and function analysis of MicroRNAs involved in exercise preconditioning-attenuating pathological cardiac hypertrophy. Int Heart J. 2018;59(5):1069-76.
Farago N, Zvara A, Varga Z, Ferdinandy P, Puskas LG. Purification of high-quality micro RNA from the heart tissue. Acta Biol Hung. 2011;62(4):413–25.
Li LL, Xue AM, Li BX, Shen YW, Li YH, Luo CL, Zhang MC, Jiang JQ, Xu ZD, Xie JH, et al. JMJD2A contributes to breast cancer progression through transcriptional repression of the tumor suppressor ARHI. Breast Cancer Res. 2014;16(3):R56.
Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20–51.
Wang X, Ha T, Liu L, Zou J, Zhang X, Kalbfleisch J, Gao X, Williams D, Li C. Increased expression of microRNA-146a decreases myocardial ischaemia/reperfusion injury. Cardiovasc Res. 2013;97(3):432–42.
Yu CH, Xu CF, Li YM. Association of MicroRNA-223 expression with hepatic ischemia/reperfusion injury in mice. Dig Dis Sci. 2009;54(11):2362–6.
Qin D, Wang X, Li Y, Yang L, Wang R, Peng J, Essandoh K, Mu X, Peng T, Han Q. And others. MicroRNA-223-5p and -3p cooperatively suppress Necroptosis in ischemic/Reperfused hearts. J Biol Chem. 2016;291(38):20247–59.
Thomas JH, Emerson RO. Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009;9:51.
Meng XY, Yu HL, Zhang WC, Wang TH, Mai X, Liu HT, Xu RC. ZFP580, a novel zinc-finger transcription factor, is involved in cardioprotection of intermittent high-altitude hypoxia against myocardial ischemia-reperfusion injury. PLoS One. 2014;9(4):e94635.
Siasos G, Tsigkou V, Tousoulis D. Circulating microRNAs as novel biomarkers in heart failure. Hell J Cardiol. 2018;59(4):215-6.
Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, Kempf T, Wollert KC, Thum T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51(5):872–5.
Acknowledgements
The authors would like to thank Dr. Jianhua Zhang and Kaifei Deng from the Academy of Forensic Science, Ministry of Justice of China for their valuable contribution to the discussion. The authors would like to also thank Dr. Wenfeng Li from the Department of Cardiology, Ganzhou People’s Hospital for collecting the human serum samples.
Funding
This study was financially supported by the Open Project of Shanghai Key Laboratory of Forensic Science (No.: KF1402) and the Open Project of Key Laboratory of Prevention and treatment of cardiovascular and cerebrovascular diseases, Ministry of Education, China (No.: XN201810), the Science and Technology Project of Jiangxi Provincial Department of Education (No.: 150952), the Science and Technology Project of Jiangxi Province (No.: 20133BBG70080) and the Key Research and Development Project of Jiangxi Province (No.: 20161BBG70082).
Author information
Authors and Affiliations
Contributions
YC performed the model establishment and microRNA array analysis. XY performed the validation assays. FY conceived, drafted and revised the manuscript. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The animal protocol was approved by the Ethical Review Board at the Gannan Medical College (No.: 2016–062) and in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication no. 85–23, revised 1996). The use of human serum samples for the purposes of basic research was also approved by the Ethical Review Board at the Gannan Medical College (No.: 2019161). Each patient was informed of the purpose of this study and showed full consent to participate into our study. The informed consents were obtained from all patients.
Consent for publication
This study did not involve human participates nor use the tissues from human sources and thus did not need patients’ consent for publication.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, Y., Ye, X. & Yan, F. MicroRNA 3113-5p is a novel marker for early cardiac ischemia/reperfusion injury. Diagn Pathol 14, 121 (2019). https://doi.org/10.1186/s13000-019-0894-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-019-0894-1