Abstract
Background
Hepatitis B virus (HBV) infection is a particular concern in human immunodeficiency virus (HIV) infected individuals. In Ethiopia, detailed clinical and virological descriptions of HBV prevailing during HIV co-infection and symptomatic liver disease patients are lacking. The aim of this study was to investigate HBV virological characteristics from Ethiopian HBV/HIV co-infected and HBV mono-infected individuals.
Methods
A total of 4105 sera from HIV positive individuals, liver disease patients, and blood donors were screened serologically for HBV. The overlapping polymerase/surface genome region of HBV from 180 infected individuals was extracted, amplified, and sequenced for genotypic analysis.
Results
The HBsAg seroprevalence was detected 43% in liver disease patients, 8.4% in blood donors, and 6.7% in HIV/HBV co-infected individuals. The occult HBV prevalence was 3.7% in HIV/HBV co-infected individuals and 2.8% in blood donors with an overall prevalence rate of 3.4%. A phylogenetic analysis showed three HBV genotypes; A (61.1%), D (38.3%) and E (0.6%). Genotype A belongs to subtypes A1 (99.1%) and A9 (0.9%), but genotype D showed heterogeneous subtypes; D2 (63.8%) followed by D4 (21.7%), D1 (8.7%), D3 (4.3%), and D10 (1.4%).
Conclusions
The HIV/HBV co-infected individuals and blood donors showed lower HBsAg seroprevalence compared to liver diseases patients. Occult HBV prevalence showed no difference between HIV/HBV co-infected and blood donor groups. This study demonstrated predominance distribution of HBV subtypes A1 and D2 in northwest Ethiopia. The observed virological characteristics could contribute for evidence-based management of viral hepatitis in Ethiopia where antiretroviral therapy guidelines do not cater for viral hepatitis screening during HIV co-infection.
Similar content being viewed by others
Background
HBV is one of the leading causes of persistent liver infections and chronic liver diseases. HBV infects an estimated 240 million people globally and shows geographical differences in its clinical consequences, seroepidemiological, and genotype distribution [1, 2]. The global HBV distribution ranges from hyperendemicity (> 8%) in areas of east Asia, Africa and the Amazon basins to intermediate (2–7%) in Europe, North America and Australia [1]. HBV is characterized by a high degree of genetic heterogeneity and classified into ten genotypes (A-J) and a number of subtypes [1]. The diversity of HBV genotypes is also related to different clinical infection patterns, liver disease severity [3], viral persistence, and response to antiviral treatments [4].
In terms of transmission routes and associated risk factors, high degrees of epidemiological similarity are common among HBV and HIV [5]. The HBV/HIV co-infected individuals are more likely to transmit hepatitis viruses, and chronic infection with HBV is a frequent clinical phenomenon [6]. In addition to the epidemiological overlap, virological and immunological interactions of HBV/HIV co-infection have also been responsible for changing the natural clinical history and the management of each virus. The introduction of highly active antiretroviral therapy (ART) has reduced the HIV/acquired immunodeficiency syndrome (AIDS) morbidity and mortality. But still, liver diseases associated with viral hepatitis became the second leading cause of death during HBV/HIV co-infection worldwide [7]. In particular, the genetic variations and clinical complications of HBV are believed to be more influenced by the presence of HIV during co-infections [5, 8], although controversy has continued over the impacts of HBV on the natural history of HIV [6, 9]. Chronic HBV/HIV co-infected patients tend to sustain higher levels of HBV viral load in their serum and affect HBV infection resolving mechanisms. More importantly, patients with HBV/HIV co-infection might be at higher risk of chronic liver disease complications, including cirrhosis and hepatocellular carcinoma [6]. As a result, the impact of viral interaction on the natural history and treatment outcomes have become increasingly important in the management of HBV/HIV co-infected individuals especially in regions where both viruses are endemic.
Moreover, treating HBV is less practiced in most African settings, although HIV is associated with a high HBV prevalence in this region [10]. Similarly, HBV was less reported in Ethiopia among liver disease patients during HIV/HBV co-infection, although considerable numbers of HBV seroepidemiological reports were available from blood donors and the general population [11]. In general, detailed clinical and virological descriptions of HBV prevailing during HIV co-infection and symptomatic liver disease patients are lacking in Ethiopia. In particular, a well-established HBV screening and clinical management is ignored in Ethiopia [12]. As a result, clinical parameters are the only means to assess liver related diseases which have limited predictive values to determine impacts of HIV during HBV co-infection. Therefore, the aim of this study was to investigate HBV serological and virological characteristics among large groups of HIV co-infected and mono-infected individuals from northwest Ethiopia.
Materials and methods
Study population
The demographic and clinical data and blood sera were collected in 2013 after ethical clearance was granted by the Institutional Ethical Review Board of University of Gondar (Ref. No: RCS/V/P/05/372/2013) and informed consent were obtained from study participants. A total of 4105 study population was enrolled from three health institutions in northwest Ethiopia; namely, University of Gondar teaching hospital, Gondar Health Center and Debretabor district hospital. The study population was recruited from three study groups; blood donors (n = 1720), liver disease patients (n = 252) and HIV carriers (n = 2133) (Table 1). The blood donors were apparently healthy, but the HIV positive group was recruited in a cohort of known HIV positive individuals who were attending ART clinics for their routine follow-ups. ART related clinical information such as ART status and eligibility, ART regimens, clinical ART toxicity assessment, World Health Organization (WHO) staging and status of opportunistic infections were recorded during sera collection. The group of liver disease patients included cases with one or more clinical manifestations of acute and chronic liver diseases patients such as jaundice, ascites, cirrhosis, hepatomegaly, hepatocellular carcinoma and/or liver complications from other infections as reported before [13].
Blood collection and serological testing
About 10 ml of venous blood was collected. The sera were screened for HBsAg using commercially available HBsAg rapid test kits (In Tec Products, INC, 332 Xinguang Road, Xiamen, China) according to manufacturers’ instruction. From those HBsAg positive sera, the HBV e antigen (HBeAg) test was done using commercially available enzyme immunoassay kits (Abbott Diagnostics, Wiesbaden, Germany).
For occult HBV (OHB) analysis, anti-HBc antibody (anti-HBcAb) test was done using Enzygnost®Anti-HBc monoclonal ELISA (Siemens Healthcare Diagnostics Products GmbH, Marburg, Germany) among 476 HBsAg negative cases. And then, the anti-HBc antibody positive sera were further tested for anti-HBs antibody using the ARCHITECT system (Abbott Diagnostics, Delkenheim, Germany). Then, the anti-HBs antibody positive and negative sera (but positive for anti-HBc antibody) were used for HBV DNA detection test and viral load determination.
The HIV-1/2 screening was done using the HIV-1/2 rapid test kits according to the Ethiopian national test algorithm adopted from WHO designed for developing countries [14]. The test algorithm used three rapid diagnostic test kits in series; HIV-(1/ + 2) Antibody Colloidal Gold (KHB, Shanghai Kehua Bio-engineering Co Ltd, China) as a screening test, followed by HIV-1/2 STAT-PAK® (Chembio HIV-1/2, Medford, New York, USA) in case of a positive screening result. In case of discordant test results with the above two kits, Unigold™ HIV (Trinity Biotech, Ireland) was used as a tiebreaker to decide the final result.
DNA extraction, PCR and sequencing
HBV DNA was extracted from 900 µl blood plasma separately using mSample preparation system nucleic acid extraction kits on the Abbott m2000sp automated sample preparation system (Abbott Molecular, Des Plaines, IL, USA). The HBV viral loads were determined using Quantitative Real Time HBV assay (lower detection rate < 10 IU/mL) on the Abbott m2000rt system (Abbott Molecular, Des Plaines, IL, USA). The HBV extraction, amplification and sequencing steps were described in detail before [15].
Briefly, for sequencing, the overlapping HBV polymerase/surface genome region (codons 52–298 nt) was amplified using Taq DNA polymerase (Promega, Madison, WI, USA) using the forward (5′AAAT TCGC AGTC CCA ACC3′) and reverse (5′GCAG CAAA GCCC AAAAG ACC3′) primers as described before [15, 16]. The amplification product was separated using 1.5% agarose gel electrophoresis, excised and purified using Wizard® SV Gel & PCR Clean-Up-System (Promega, Mannheim, Germany). Clean HBV DNA PCR products were subjected to direct sequencing of both forward and reverse strands using BigDye Terminator Cycle Sequencing Ready Reaction kit on the ABI Prism 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
After the nucleotide sequences were manually edited and assembled using the Geneious® software version 6.1.4 (http://www.geneious.com), genotyping analysis was done using MEGA 6 (http://www.megasoftware.net). For phylogenetic analysis, HBV genotypes reference sequences were retrieved from the HBV database (https://hbvdb.ibcp.fr/HBVdb/HBVdbNomenclature?nomenclature=table). In addition, a set of homologous sequences was retrieved from GenBank Basic Local Alignment Search Tool (BLAST). The phylogenetic tree was constructed using the Neighbor-Joining method, and the genetic distances were computed using the Kimura 2-parameter method (http://www.megasoftware.net). To confirm the reliability of the phylogenetic tree, a bootstrap resampling test was carried out 1000 times. The HBV nucleotide sequences used in this study were registered to the GenBank/EMBL/DDBJ databases under the accession numbers KT367571-KT367731 and OL630698-OL630716.
Statistical analysis
Mann–Whitney non-parametric and Chi-square tests were used when appropriate during analysis. Graph Pad Prism version 5.01, 2007 was used for statistical analysis. A P value < 0.05 was considered to be statistically significant.
Results
Serological and viral load characteristics of HBsAg positives
Details of demographic and clinical characteristics of HBsAg-HIV co-infected study participants are presented in Table 2. The seroprevalence of HBsAg among HIV co-infected individuals; 6.7% (143) was significantly lower than in blood donors; 8.4% (145) (p = 0.04) and liver disease patients; 43.3% (109) (p < 0.001) (Fig. 1A). In contrast, the HBeAg frequency was significantly higher among HIV positives (44.6% (25), p = 0.02) and liver disease patients (41.8% (23), p = 0.04) than blood donors; 21.7% (10) (Fig. 1B).
The HBV viremia rate showed significant differences with a distribution frequency of 55.5% (71/128) in HIV/HBV co-infected individuals, 79.6% (82/103) in blood donors and 89.9% (98/109) in liver disease patients (Table 1). Among HIV/HBV co-infection, HBV viraemic rate was significantly lower among ART experienced; 47.5% (38/80) than ART naives; 68.8% (33/48) (p = 0.02). The overall median (Interquartile range; IQR) viral load levels were also compared in each study group and showed significant highest difference in the HIV/HBV co-infected individuals than blood donors and liver disease patients (Fig. 2). Similarly, among the HBV sequenced sera, HIV/HBV co-infected individuals; 6.4 log IU/ml (3.1–8.3) showed higher viral load levels than blood donors; 3.3 log IU/ml (2.7–3.8) and liver disease patients; 5.0 log IU/ml (3.9–6.8) (Fig. 2). However, the median (IQR) HBV viral load levels showed no significant differences between ART experienced; 4.6 log IU/ml (3.1–8.4) and ART navies; 6.5 log IU/ml (2.9–7.9) (p = 0.89).
Serological and viral load characteristics of occult HBV
The anti-HBcAb positivity was 57.0% (171/300) and 50.6% (89/176), respectively among the HBV/HIV co-infected individuals and blood donors, which revealed no statistical significance (p = 0.21) (Fig. 3A). The anti-HBs negative frequency among the anti-HBcAb positives showed no significant difference between HIV co-infected individuals; 25.7% (77) and blood donors; 18.8% (33) (p = 0.11) (Fig. 3B). Among the anti-HBc antibody positive cases, 14.3% (11/77) from HIV co-infected and 15.2% (5/33) from blood donors showed HBV DNA positivity (Table 3). The overall seroprevalence of OHB infection (HBsAg negative but DNA positive) was 3.4%. However, the seroprevalence became 3.7% (11/300) and 2.8% (5/176) in the HIV co-infected individuals and blood donors, respectively. The viral load for the OHB infection ranges from < 1 to 3.69 log IU/ml.
The seroprevalence comparisons of anti-HBcAb positive (A) and anti-HBs negative (B) markers among HBsAg negative blood donors and HIV co-infected groups. The anti-HBc antibody and anti-HBs tests were not considered among the HBsAg negative sera of liver disease patient due to a relative low study population representation than blood donors and HIV co-infected groups
Genetic diversity of HBV
An overall of 180 HBV DNA positive sera (177 from HBsAg positive cases and 3 from OHB infection) was sequenced and genotyped (Tables 1 and 3) with a representation of 36.1% (65) HBV/HIV co-infected, 31.6% (56) blood donors, and 33.3% (59) liver disease patients. The genotypic analysis revealed three HBV genotypes; 61.1% (110) genotype A, 38.3% (69) genotype D and 0.6% (1) genotype E. Further sub-genotypic analysis showed genotype A sub-typed as A1, 99.1% (109) and A9, 0.9% (1). Among genotype D, the subtypes distribution showed 63.8% (44) subtype D2, followed by 21.7% (15) subtype D4, 8.7% (6) subtype D1, 4.3% (3) subtype D3, and 1.4% (1) subtype D10.
Genotype A showed no significant (p > 0.05) difference distribution among HIV co-infected; 33.6% (n = 37), blood donors; 33.6% (n = 37), and liver disease patients; 32.7% (n = 36). Similarly, genotype D was also distributed with 39.1% (n = 27), 27.5% (n = 19) and 33.3% (n = 23) among the above respective groups (p > 0.05).
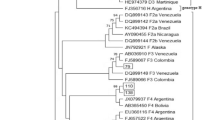
The phylogenetic analysis revealed that the majority of genotype A clustered within the African subtype, but some were clustered to the Asian, Americas (Haiti) and Australia subtype. Genotype D was commonly clustered to East African, North African, Middle East and European strains. The single isolate genotype E showed the highest homology with African strains (Fig. 4).
Phylogenetic tree analysis of the HBV sequences isolated among different population category from Ethiopia. The phylogenetic tree was constructed based on the HBV pol/S genome region (codons 52-298). The HBV isolates (denoted by ETH initials) were analyzed with respect to reference sequences retrieved from the GenBank, which are designated by their respective accession numbers (marked as ●) along HBV genotypes and country of origins [AUS-Australia, EGY-Egypt, ET-Ethiopia, GHN-Ghana HAI-Haiti, IND-India, ITA-Italy, KEN-Kenya, OMN-Oman, POL-Poland, RUS-Russia, RWN-Rwanda, SA-S. Africa, SOM-Somalia, SUN-Sudan, TAJ-Tajikistan, TAN-Tanzania, TUN-Tunisia, TUR-Turkey, UAE-United Arab Emirates, UGA-Uganda, UK-United Kingdom]. The genome sequence of the Woolly monkey hepatitis B virus (GenBank AY226578; marked as ♦) was utilized as an out-group for the rooted tree analysis
Discussion
According to HBV epidemiological geographical variations [17], Ethiopia can be categorized under the high intermediate (5–7%) to hyperendemicity (≥ 8%) HBV prevalence. The HBsAg prevalence from 6.7% in HIV/HBV infected individuals and 8.4% in blood donors to 43.3% in liver disease patients was recorded in this study. The HBV prevalence from blood donors was in accordance with reports from other African countries [18,19,20]. Similarly, HBV prevalence in the liver disease patients was comparable to similar reports from symptomatic patients in Africa [21]. The relative lower prevalence of HBV in HIV infected individuals to blood donors and liver disease patients in this study and to other similar studies in Africa (9.0–16.9%) [10, 22, 23] might be partly associated by ART drugs like lamivudine and tenofovir used for HIV therapy during co-infection [24] since the majority of (70.4%) HIV co-infected cases screened for HBV in this study were drug experienced under different ART regimens. Nevertheless, the 6.7% HBV in the current study was within the range of previously reported HBV prevalence (3.7–7.4%) among HIV co-infected individuals in Ethiopia [11]. Moreover, the HBV/HIV co-infected population in the current study was also characterized by a presence of classical and putative immune escape HBsAg gene variants [16] that down-regulate HBsAg-antibody binding surface proteins which usually associated to a high chance of HBV false negativity due low HBsAg [25]. But still, within HBV tested positive cases, the HBV viraemic rate is still high among HIV patients despite ART therapy. This could be associated to the HIV therapy practiced for many years in the study area without testing patients for their HBV status which was responsible for high rate of a combination of multi-drug resistance gene variability observed and reported before [16].
In contrast to lower HBsAg detection rate, however, an almost two times higher HBeAg frequency coupled with significantly higher median viral load levels were observed in these HBV/HIV-infected individuals. Nevertheless, the observation of the significantly higher rate of anti-HBcAb in the HBsAg negative HIV infected study group showed a relatively higher rate of past infection during HIV co-infection. However, the anti-HBs status that showed immunity following past infection were nearly similar from HIV infected and mono-infected groups. This could support the fact that the chance of HBV exposure might not be different among the study groups in the study setting. In Ethiopia, HBV vaccination was introduced in 2007 administered for new born and the current adult study population had no vaccine exposure. Reports also linked to HIV infection to ‘sero-silent’ OHB infections, which presents serious problems for diagnosis, prevention, and control [22]. In this HIV/HBV co-infected individuals of the current study, 3.7% of OHB infection was detected which highlights the need for integrating OHB screening for proper management of liver diseases in HIV co-infected patients.
In this study, the phylogenetic analysis revealed circulation of three genotypes; genotype A, genotype D and genotype E. Overall, genotype A was predominant and both genotypes A and D were the most divergent as they clustered to clades of African, Asian, Americas, Europe, Middle East and Oceania clades. In East Africa, where Ethiopia belongs to, the predominance of genotype A was found in Somalia to the east [26] and in Kenya [27] to the south. However, in neighboring Sudan to the west of Ethiopia, where geographical proximity is close and large influx of population movement through trade and migration are common to the current study settings in northwest Ethiopia, the identified genotypes showed a different distribution pattern. Unlike in Ethiopia where genotype A was predominant, in Sudan genotypes D and E were predominant [19]. Only one isolate with genotype E was identified in this study, but of course with the highest homology to the Sudanese strains.
Although genotype A is known as one of the heterogeneous HBV genotypes [28], a predominance of subtype A1 was identified in this study. This was actually in accordance with most African-derived isolates sequenced so far belonging to subtype A1. The predominance of subtype A1 and the phylogenetic similarity of the majority of the isolates with the neighboring Somalian and other East African isolates also support the notation that this was evolved within the indigenous population of some African countries and had a longer natural history in Africa than other subtypes [29].
Unlike genotype A, subtypes of genotype D were heterogeneous (D1, D2, D3, D4 and D10) in this study population. In fact, subtypes D1 to D5 are known for their wider global distribution among so far described nine subtypes (D1-D9) [1, 28, 30]. In this study, subtype D2 (61.1%) followed by D4 (21.7%) were dominant, although a recent report from blood donors in Ethiopia reported D4 as the least (2.5%) subtype and also failed to identify the subtype D3 [31]. Nevertheless, the subtype D3 was exclusively found among liver disease patients in this study. Previous studies showed subtype D1 was predominant in the Eastern part of African countries [32], despite the current study and an earlier report from neighboring Somalia [29] reported D2 and D4 were predominant. As a result, these subtypes might be speculated to be indigenous in Ethiopia and Somalia. This is because the nearby Middle East and northern African regions [1] were known for the subtype D1, but the frequency of this subtype was very low (8.7%) in the current study. Moreover, the geographic proximity of countries, mostly associated to subtypes D2, D3, and D4 (South Africa, Alaska, south to north-eastern Europe and Oceania) [1, 33] are far distant from east Africa, although few isolates in the current study had a phylogenetic relationship to distant isolates such as from Oman, UK and Italy. Actually, given to the polymerase/surface gene sequences used in the phylogenetic analysis in this study, such conclusion might be partial unless full genome sequences are compared since HIV co-infection is known to be responsible for HBV variability and high rate of recombination [34]. For instance, a recently described new D8 and D9 subtypes from Niger and India, respectively recognized as recombinant forms of genotype D with genotype E [35] and genotype C [36]. A novel hepatitis B virus subtype D10 was also reported circulating in Ethiopia [37], but only a single isolate was identified with this subtype in the current study. Overall, this study showed subtypes of genotype D were heterogeneous in northwest Ethiopia.
Conclusions
In summary, relative to the blood donors, HIV/HBV co-infected group was characterized by lower HBV seroprevalence, but showed higher HBeAg detection rate and median viral load levels. Moreover, the anti-HBcAb, which showed past HBV infection was significantly higher in HIV co-infected group but showed no difference to anti-HBs (a HBV marker for immunity to past infection) as well as OHB infection. The phylogenetic analysis showed circulation of heterogeneous HBV subtypes (A1, A9, D1, D2, D3, D4, D10 and E) with the predominance of subtypes A1 and D2 in Ethiopia. The HBV subtypes showed the most divergent isolates clustered to both African and non-African clades. This is the first HBV genetic diversity study among HIV co-infection and symptomatic liver disease patients in Ethiopia. These data will have significant public health implications to understand further clinical and virological interplay of HBV during HIV co-infections in this country where viral hepatitis management was totally ignored and HIV antiretroviral guidelines do not even cater for viral hepatitis during HIV co-infection.
Availability of data and materials
The HBV nucleotide sequences used in this study were registered to the GenBank/EMBL/DDBJ databases under the accession numbers KT367571-KT367731 and OL630698-OL630716.
Abbreviations
- AIDS:
-
Acquired immunodeficiency syndrome
- Anti-HBs:
-
Anti- Hepatitis B surface
- ART:
-
Active antiretroviral therapy
- BLAST:
-
Basic Local Alignment Search Tool
- ELISA:
-
Enzyme-linked immunosorbent assay
- HBcAb:
-
Hepatitis B core antibody
- HBsAg:
-
Hepatitis B surface antigen
- HBV:
-
Hepatitis B virus
- HIV:
-
Human immunodeficiency virus
- MEGA:
-
Molecular Evolutionary Genetics Analysis
- PCR:
-
Polymerase chain reaction
- OHB:
-
Occult Hepatitis B virus
- WHO:
-
World Health Organization
References
Zehender G, Ebranati E, Gabanelli E, Sorrentino C, Lo Presti A, et al. Enigmatic origin of hepatitis B virus: an ancient travelling companion or a recent encounter. World J Gastroenterol. 2014;20(24):7622–34.
Ward H, Tang L, Poonia B, Kottilil S. Treatment of hepatitis B virus: an update. Future Microbiol. 2016;11(12):1581–97.
Tsubota A, Arase Y, Ren F, Tanaka H, Ikeda K, et al. Genotype may correlate with liver carcinogenesis and tumor characteristics in cirrhotic patients infected with hepatitis B virus subtype adw. J Med Virol. 2001;65(2):257–65.
Rajoriya N, Combet C, Zoulim F, Janssen HLA. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualized approach? J Hepatol. 2017;67(6):1281–97.
Matthews PC, Geretti AM, Goulder, Philip JR, Klenerman P. Epidemiology and impact of HIV co-infection with hepatitis B and hepatitis C viruses in Sub-Saharan Africa. J Clin Virol 2014; 61(1):20–33.
Singh KP, Crane M, Audsley J, Avihingsanon A, Sasadeusz J, Lewin SR. HIV-hepatitis B virus co-infection: epidemiology, pathogenesis, and treatment. AIDS. 2017;31(15):2035–52.
Dharel N, Sterling RK. Hepatitis B Virus-HIV coinfection: forgotten but not gone. Gastroenterol Hepatol. 2014;10(12):780–8.
Weber R, Ruppik M, Rickenbach M, Spoerri A, Furrer H, et al. Decreasing mortality and changing patterns of causes of death in the Swiss HIV Cohort Study. HIV Med. 2013;14(4):195–207.
Mu Y, Yu Y, Yue X, Musarat I, Gong R, Zhu C, Liu Y, Liu F, Zhu Y, Wu J. The X protein of HBV induces HIV-1 long terminal repeat transcription by enhancing the binding of C/EBPβ and CREB1/2 regulatory proteins to the long terminal repeat of HIV-1. Virus Res. 2011;156(1–2):81–90.
Barth RE, Huijgen Q, Taljaard J, Hoepelman, Andy IM. Hepatitis B/C and HIV in sub-Saharan Africa: an association between highly prevalent infectious diseases. A systematic review and meta-analysis. IJID 2010;14(12): e1024–31.
Belyhun Y, Maier M, Mulu A, Diro E, Liebert UG. Hepatitis viruses in Ethiopia: a systematic review and meta-analysis. BMC Infect Dis. 2016;16(1):761.
Federal Democratic Republic of Ethiopia Ministry of Health: National guidelines for comprehensive HIV prevention, care and treatment. Addis Ababa, 2018, Ethiopia.
Belyhun Y, Liebert UG, Maier M. Analysis of HBV basal core promoter/precore gene variability in patients with HBV drug resistance and HIV co-infection in Northwest Ethiopia. PloS One. 2018;13(2):e0191970.
Shanks L, Siddiqui M, Kliescikova J, Pearce N, Ariti C, et al. Evaluation of HIV testing algorithms in Ethiopia. The role of the tie-breaker algorithm and weakly reacting test lines in contributing to a high rate of false positive HIV diagnoses. BMC Infect Dis. 2015;15(1):39.
Maier M, Liebert UG, Wittekind C, Kaiser T, Berg T, Wiegand J. Clinical relevance of minimal residual viremia during long-term therapy with nucleos(t)ide analogues in patients with chronic hepatitis B. PLoS ONE. 2013;8(6):e67481.
Belyhun Y, Maier M, Liebert UG. HIV therapy with unknown HBV status is responsible for higher rate of HBV genome variability in Ethiopia. Antivir Ther. 2017;22(2):97–111.
Otta JJ, Stevensa GA, Groegerb J, Wiersmaa ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–9.
Jayaraman S, Chalabi Z, Perel P, Guerriero C, Roberts I. The risk of transfusion-transmitted infections in sub-Saharan Africa. Transfusion. 2010;50(2):433–42.
Mahgoub S, Candotti D, El Ekiaby M, Allain J. Hepatitis B virus (HBV) infection and recombination between HBV genotypes D and E in asymptomatic blood donors from Khartoum. Sudan J Clin Microbiol. 2011;49(1):298–306.
Musa BM, Samaila AA, Femi OL, Borodo MM, Bussell S. Prevalence of hepatitis B virus infection in Nigeria, 2000–2013. A systematic review and meta-analysis. Niger J Clin Pract. 2015;18(2):163.
Lemoine M, Thursz MR. Battlefield against hepatitis B infection and HCC in Africa. J Hepatol. 2017;66(3):645–54.
Burnett RJ, François G, Kew MC, Leroux-Roels G, Meheus A, Hoosen AA, Mphahlele MJ. Hepatitis B virus and human immunodeficiency virus co-infection in sub-Saharan Africa: a call for further investigation. Liver Int. 2005;25(2):201–13.
Franzeck FC, Ngwale R, Msongole B, Hamisi M, Abdul O, et al. Viral hepatitis and rapid diagnostic test-based screening for HBsAg in HIV-infected patients in rural Tanzania. PLoS ONE. 2013;8(3):e58468.
Núñez M, Puoti M, Camino N, Soriano V. Treatment of chronic hepatitis B in the human immunodeficiency virus-infected patient: present and future. Clin Infect Dis. 2003;37(12):1678–85.
Lazarevic I. Clinical implications of hepatitis B virus mutations. Recent Adv WJG. 2014;20(24):7653.
Kramvis A, Kew MC. Molecular characterization of subgenotype A1 (subgroup Aa) of hepatitis B virus. Hepatol Res. 2007;37(s1):S27-32.
Mwangi J, Nganga Z, Songok E, Kinyua J, Lagat N, et al. Molecular genetic diversity of hepatitis B virus in Kenya. Intervirol. 2008;51(6):417–21.
Kramvis A, Kew MC. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol Res. 2007;37(s1):S9–19.
Andernach IE, Nolte C, Pape JW, Muller CP. Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. EID. 2009;15(8):1222–8.
Chandra PK, Biswas A, Datta S, Banerjee A, Panigrahi R, et al. Subgenotypes of hepatitis B virus genotype D (D1, D2, D3 and D5) in India: differential pattern of mutations, liver injury and occult HBV infection. J Viral Hepat. 2009;16(10):749–56.
Hundie GB, Raj VS, Gebre Michael D, Pas SD, Osterhaus A, et al. Molecular epidemiology and genetic diversity of hepatitis B virus in Ethiopia. J Med Virol. 2015;88(6):1035–43.
Saudy N, Sugauchi F, Tanaka Y, Suzuki S, Aal AA, et al. Genotypes and phylogenetic characterization of hepatitis B and delta viruses in Egypt. J Med Virol. 2003;70(4):529–36.
Zehender G, Ebranati E, Gabanelli E, Shkjezi R, Lai A, et al. Spatial and temporal dynamics of hepatitis B Virus D genotype in Europe and the Mediterranean Basin. PLoS ONE. 2012;7(5):e37198.
Martin CM, Welge JA, Blackard JT. Hepatitis B virus (HBV) X gene diversity and evidence of recombination in HBV/HIV co-infected persons. J Med Virol. 2011;83(7):1142–50.
Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, et al. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91(Pt 6):1609–20.
Ghosh S, Banerjee P, Deny P, Mondal RK, Nandi M, et al. New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India. J Viral Hepat. 2013;20(3):209–18.
Hundie GB, Stalin Raj V, Gebre Michael D, Pas SD, Koopmans MP, Osterhaus AD, et al. A novel hepatitis B virus subgenotype D10 circulating in Ethiopia. J Viral Hepat. 2017;24(2):163–73.
Acknowledgements
The authors would like to thank study participants and staffs at Serology Section, Blood Bank, HIV Testing and Voluntary Counseling Center, and HIV/AIDS ART Clinic of University of Gondar Hospital, Debretabour Hospital, and Gondar Poly Health Center in Ethiopia. We are grateful to Banchamlak Asmare, Getnet Ayelew, Amare Kiflie, Mengistu Endris, Prof. Feleke Moges, Melashu Balew and Zuriayash Alemu for their assistance during data collection. Expert technical assistance by Dr. Andargachew Mulu, Susanne Hummel and Kathleen Zielinski is acknowledged.
Funding
This study was supported partly by the German Academic Exchange Service (DAAD) Friends and Sponsors of Leipzig University, and an award of Emeritus Prof. Fusao Ota's Scholarship for Young Ethiopian Researcher, 2010. http://web.me.com/fusao_ota/Japan-Ethiopia/toppage.Eng.html.
Author information
Authors and Affiliations
Contributions
YB, UGL and MM equally contributed to this study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was granted from the Institutional Ethical Review Board of University of Gondar (Ref. No: RCS/V/P/05/372/2013) and permission was obtained from the University of Gondar teaching Hospital, Debretabor Hospital, and Gondar Poly Health Centre, Ethiopia (Ref. No: CMHS/08/30/2013). Informed consent from each study participant was also obtained before data collection.
Consent for publication
Not applicable.
Competing interests
The authors declare that we do not have any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Belyhun, Y., Liebert, U.G. & Maier, M. Molecular epidemiology of hepatitis B virus among HIV co-infected and mono-infected cohorts in Northwest Ethiopia. Virol J 19, 53 (2022). https://doi.org/10.1186/s12985-022-01774-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-022-01774-6