Abstract
Background
Cucurbit chlorotic yellows virus (CCYV), a bipartite crinivirus, causes chlorotic leaf spots and yellowing symptoms on cucurbit leaves. We previously developed an infectious clone of CCYV. Limited work has been conducted on the construction of a crinivirus green fluorescence protein (GFP) expression vector to date.
Finding
We constructed a CCYV GFP expression vector using the “add a gene” strategy based on CCYV RNA2 cDNA constrcut. Three resultant clones, pCCYVGFPSGC, pCCYVGFPCGC, and pCCYVGFPCGS, were constructed with different promoters used to initiate GFP and CP expression. At 25 dpi GFP fluorescence was detectable not only in leaf veins but also in the surrounding cells. pCCYVGFPCGC-infected cucumber leaves exhibited cell spread at 25 dpi, whereas pCCYVGFPSGC and pCCYVGFPCGS were mainly found in single cells. Further observation of pCCYVGFPCGC GFP expression at 30 dpi, 40 dpi, and 50 dpi showed phloem-limited localization in the systemic leaves.
Conclusions
We developed of a CCYV GFP expression vector that will be useful for further study of CCYV movement in cucurbits.
Similar content being viewed by others
Cucurbit chlorotic yellows virus (CCYV), a recently discovered cucurbit-infecting crinivirus in the family Closteroviridae [1,2,3,4,5,6,7], is among the largest single-strand positive-sense RNA viruses [8]. The bipartite RNA genome comprises a 8607 nucleotide [nt] RNA1 and a 8041-nt RNA2. Recent studies on CCYV [9,10,11,12] have been hampered due to lack of reverse genetic tools. The construction of full-length infectious cDNA clones will facilitate the investigation of viral determinants of virus replication and movement, as well as the interactions between viral proteins and host factors. Our previous study developed two sets of full-length CCYV cDNA clones under the control of the T7 RNA polymerase promoter and 35S promoter [13].
Virus-based vectors are useful tools for the study of plant molecular biology. A number of plant virus vectors have been developed to express heterologous genes of interest in plants [14,15,16,17,18]. Only LIYV has been reported to construct a green fluorescence protein (GFP) vector for crinivirus using the ORF fusion strategy. Part of LIYV RNA1 encoded P34 protein was replaced by a GFP gene and further observation of Nicotiana benthamiana (Nb) leaves agro-infiltrated by the resultant clone displayed occasional single cells green fluorescence. Limited GFP expression has been observed in Nicotiana benthamiana plants, but in no other hosts. Cucumber is an economically important cultivated plant, and the available GFP expression vector of cucurbit viruses is limited [19]. In this study, we constructed a CCYV GFP expression vector using the “add a gene” strategy with an extra subgenomic RNA and a foreign GFP protein, according to a previous study of the closterovirus Citrus tristeza virus [20], which suggested that this strategy would be more appropriate than ORF substitution or ORF fusion. We examined the systemic leaves of cucumber plants regarding the resulting GFP fluorescence.
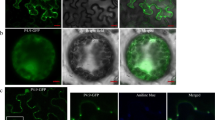
We chose to insert gfp ORF before the CP gene, because the CP subgenomic controller elements (CEs) are more suitable than other CEs [20,21,22,23]. The full-length CCYV RNA2 clone was modified to accommodate the gfp gene immediately upstream of the CP open reading frame via overlapping polymerase chain reaction (PCR). Sweet potato chlorotic stunt virus (SPCSV) CP CEs were used to direct the expression of the CCYV CP gene or GFP. The strategy used to construct the full-length cDNA clones of RNA1 and RNA2 is outlined in Fig. 1. The resulting clones were named pCCYVGFPSGC, pCCYVGFPCGC, and pCCYVGFPCGS. To construct the clone pCCYVGFPCGS, four overlapping fragmens were amplified using the primer pairs showed in Table 1 and the recombinant fragment were obtained using overlap PCR. Restriction enzymes MluI and NruI were used to insert the fragment into the pCBCCYVRNA2 to acquire the GFP-tagged vector pCCYVGFPCGS. Based on pCCYVGFPCGS, a fragment from 4811 to 5193 nt covering CCYVCP CEs and partial CP sequence was amplified using primer pair CCYVCcgccF2F/CCYVCcgscF2R. pCCYVGFPCGS and the fragment were digested by PacI and NruI to acquire the recombinant clone pCCYVGFPCGC. pCCYVGFPSGC vector was cloned based on pCCYVGFPCGC. Three overlapping fragments were amplified using primer pairs CCYVcgscF1F/CCYVsgccF1R, CCYVsgccSPCSVCPpF/CCYVsgccSPCSVCPpR, and CCYVsgccGFPF/CCYVcgscF2R, and overlap PCR was used to ligate the fragments and inserted into pCBCCYVRNA2 vector using restriction enzymes MluI and NruI. The resultant clones were transformed into Agrobacterium tumefaciens strain GV3101 for agroinfiltration. At 25 days after agroinfiltration of pCCYVGFP with pCBCCYVRNA1 and pCBP1/HC-Pro in C. sativus, the adaxial side of leaf was used to observe the GFP fluorescence using epifluorescence microscope (Nikon ECLIPSE Ti-S) with the excitation of 460-480 nm and emission wavelength of 500-540 nm, respectively (Fig. 2a). GFP fluorescence was detectable not only in leaf veins, but also in the surrounding cells. This result is consistent with that of a previous study of CCYV localization in leaf laminae using immunoblots [9]. We observed that pCCYVGFPCGC-inoculated cucumber leaves exhibited cell spread at 25 dpi, whereas the other two constructs were mainly found in single cells (Fig. 2b). The number of pCCYVGFPCGC fluorescent cells was significantly higher than that of pCCYVGFPCGS fluorescent cells. Further observation of pCCYVGFPCGC at 30 dpi showed that GFP was mainly vein-limited in cucumber systemic leaves (Fig. 3). Time course observation of pCCYVGFPCGC GFP fluorescence showed that at 40 dpi the GFP fluorescence was mainly focused in the second systemic leaf and at 50 dpi the fluorescence was mainly focused in the main vein of systemic leafs showing the most fluorescence in the third systemic leaf (Fig. 3).
Schematic representation of the construction of a green fluorescent protein (GFP) expression vector of Cucurbit chlorotic yellows virus (CCYV). GFP was inserted immediately upstream of the CP gene. Three different combinations of CCYV CP controller elements (CEs) and Sweet potato chlorotic stunt virus (SPCSV) CP CEs are shown. Red square indicates CEs of SPCSVCP; black square indicates CEs of CCYVCP. Grey square indicates the gfp gene
CCYV GFP expression in the systemic leaves of cucumber at 25 dpi. a From left to right: GFP expression in the leaf vein, GFP expression in a single cell beside the leaf vein, and GFP expression in the surrounding cells. Bars represent 200 μm. b Counting expression spots of the different constructs on systemic leaves. We selected 10 different visual fields for counting. Dark grey bars indicates the single fluorescent cells and light grey bars indicates the multiple cells showing fluorescence. Sgc, cgc, cgs represents the three different strategies used for GFP expression. * indicates significant difference with P value<0.05
As previously reported for the closterovirus CTV, a heterologous BYV sgRNA CE to control GFP expression was stable and worked best of the stratefies examined [20]. The closterovirus beet yellow stunt virus (BYSV) 248 bp upstream of the CP coding region was chosen to direct the expression of BYV CP, and the original CP promoter was used to direct GFP expression [24]. We chose a 201-bp CE sequence upstream of the SPCSV CP coding region to test the heterologous effect, and a 131-bp CE of CCYV CP to test the homologous effect. According to our results, the homologous sequence repeats in the vector promoted the GFP expression of CCYV, whereas when we used the CE of SPCSV CP to direct expression of CCYV CP or GFP, GFP expression was delayed. Hence, pCCYVGFPCGC can be used as a candidate for further study of GFP expression. Here we tested the insertion of GFP before the CP coding region. Other positions could be tested further to see if the other position would work better.
Abbreviations
- CCYV:
-
Cucurbit chlorotic yellows virus
- CEs:
-
controller elements
- GFP :
-
green fluorescent protein
- SPCSV:
-
Sweet potato chlorotic stunt virus
References
Abrahamian P, Sobh H, Abou-Jawdah YA. First report of Cucurbit chlorotic yellows virus on cucumber in Lebanon. Plant Dis. 2012;96:1704.
Gu QS, Liu YH, Wang YH, Huangfu WG, Gu HF, Xu L, Song FM, Brown JK. First report of Cucurbit chlorotic yellows virus in cucumber, melon, and watermelon in China. Plant Dis. 2011;95:1168.
Huang LH, Tseng HH, Li JT, Chen TC. First report of Cucurbit chlorotic yellows virus infecting cucurbits in Taiwan. Plant Dis. 2010;94:1168.
Keshawarz T, Shams-Bakhsh M, Izadpanah K, Malboobi MA. Occurrence and genome analysis of Cucurbit chlorotic yellows virus in Iran. J Phytopathol. 2014;162:523–6.
Okuda M, Okazaki S, Yanasaki S, Okuda S, Sugiyama M. Host range and complete genome sequence of Cucurbit chlorotic yellows virus, a new member of the genus crinivirus. Phytopathology. 2010;100:560–6.
Orfanidou C, Maliogka VI, Katis NI. First report of Cucurbit chlorotic yellows virus in cucumber, melon, and watermelon in Greece. Plant Dis. 2014;98:1446–7.
Hamed K, Menzel W, Dafalla G, Gadelseed AMA, Winter S. First report of Cucurbit chlorotic yellows virus infecting muskmelon and cucumber in Sudan. Plant Dis. 2011;95:1321.1.
Martelli IM, Agranovsky AA, Bar-Joseph M, Boscia D, Candresse T, Coutts RHA, et al. Closteroviridae. In: Andrew MQ, King MJA, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy classification and nomenclature of viruses ninth report of the international committee taxonomy of viruses. USA: Elsevier Inc.; 2012. p. 987–1001.
Kubota K, Usugi T, Tsuda S. Production of antiserum and immunodetection of Cucurbit chlorotic yellows virus, a novel whitefly-transmitted crinivirus. J Gen Plant Pathol. 2011;77:116–20.
Okuda S, Okuda M, Sugiyama M, Sakata Y, Takeshita M, Iwai H. Resistance in melon to Cucurbit chlorotic yellows virus, a whitefly-transmitted crinivirus. Eur J Plant Pathol. 2013;135:313–21.
Wang Z, Gu Q, Sun H, Li H, Sun B, Liang X, et al. One-step reverse transcription loop mediated isothermal amplification assay for sensitive and rapid detection of Cucurbit chlorotic yellows virus. J Virol Methods. 2014;195:63–6.
Wang ZY, Wang YZ, Sun H, Gu QS, Li HL, Sun BJ, et al. Two proteins of Cucurbit chlorotic yellows virus, P59 and P9, are self-interacting. Virus Genes. 2015;51:152–5.
Shi Y, Chen L, Sun B, Sun X, Wang Z, Gu Q, et al. Infectious clones of the crinivirus Cucurbit chlorotic yellows virus are competent for plant systemic infection and vector transmission. J Gen Virol. 2016;97(6):1458–61.
Baulcombe DC, Chapman S, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7(6):1045–53.
Toth RL, Chapman S, Carr F, Santa Cruz S. A novel strategy for the expression of foreign genes from plant virus vectors. FEBS Lett. 2001;489(2-3):215–9.
Liu Z, Kearney CM. A tobamovirus expression vector for agroinfection of legumes and Nicotiana. J Biotechnol. 2010;147(3-4):151–9.
Shivprasad S, Pogue GP, Lewandowski DJ, Hidalgo J, Donson J, Grill LK, et al. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology. 1999;255(2):312–23.
Canizares MC, Lomonossoff GP, Nicholson L. Development of cowpea mosaic virus-based vectors for the production of vaccines in plants. Expert Rev Vaccines. 2005;4(5):687–97.
Rhee S-J, Jang YJ, Lee GP. Identification of the subgenomic promoter of the coat protein gene of cucumber fruit mottle mosaic virus and development of a heterologous expression vector. Arch Virol. 2016;161(6):1527–38.
Folimonov AS, Folimonova SY, Bar-Joseph M, Dawson WO. A stable RNA virus-based vector for citrus trees. Virology. 2007;368(1):205–16.
Ayllon MA, Gowda S, Satyanarayana T, Dawson WO. cis-acting elements at opposite ends of the Citrus tristeza virus genome differ in initiation and termination of subgenomic RNAs. Virology. 2004;322(1):41–50.
Gowda S, Satyanarayana T, Davis CL, Navas-Castillo J, Albiach-Marti MR, Mawassi M, et al. The p20 gene product of Citrus tristeza virus accumulates in the amorphous inclusion bodies. Virology. 2000;274(2):246–54.
Kurth EG, Peremyslov VV, Prokhnevsky AI, Kasschau KD, Miller M, Carrington JC, et al. Virus-derived gene expression and RNA interference vector for grapevine. J Virol. 2012;86(11):6002–9.
Peremyslov VV, Hagiwara Y, Dolja VV. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc Natl Acad Sci U S A. 1999;96(26):14771–6.
Acknowledgements
We thank Dr. Fengmin Yan for helpful comments on the preparation of this manuscript.
Funding
Financial support was provided by National Natural Science Foundation of China (31701944).
Author information
Authors and Affiliations
Contributions
YS and ZW designed the experiment. YW performed the experiment and data analysis. YS and ZW wrote paper. QG, LC, HL, BS, XH were involved in discussion. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors consent to publish.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wei, Y., Han, X., Wang, Z. et al. Development of a GFP expression vector for Cucurbit chlorotic yellows virus. Virol J 15, 93 (2018). https://doi.org/10.1186/s12985-018-1004-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-018-1004-9