Abstract
Background
Abacavir and rilpivirine are alternative antiretroviral drugs for treatment-naïve HIV-infected patients. However, both drugs are only recommended for the patients who have pre-treatment HIV RNA <100,000 copies/mL. In resource-limited settings, pre-treatment HIV RNA is not routinely performed and not widely available. The aims of this study are to determine factors associated with pre-treatment HIV RNA <100,000 copies/mL and to construct a model to predict this outcome.
Methods
HIV-infected adults enrolled in the TREAT Asia HIV Observational Database were eligible if they had an HIV RNA measurement documented at the time of ART initiation. The dataset was randomly split into a derivation data set (75% of patients) and a validation data set (25%). Factors associated with pre-treatment HIV RNA <100,000 copies/mL were evaluated by logistic regression adjusted for study site. A prediction model and prediction scores were created.
Results
A total of 2592 patients were enrolled for the analysis. Median [interquartile range (IQR)] age was 35.8 (29.9–42.5) years; CD4 count was 147 (50–248) cells/mm3; and pre-treatment HIV RNA was 100,000 (34,045–301,075) copies/mL. Factors associated with pre-treatment HIV RNA <100,000 copies/mL were age <30 years [OR 1.40 vs. 41–50 years; 95% confidence interval (CI) 1.10–1.80, p = 0.01], body mass index >30 kg/m2 (OR 2.4 vs. <18.5 kg/m2; 95% CI 1.1–5.1, p = 0.02), anemia (OR 1.70; 95% CI 1.40–2.10, p < 0.01), CD4 count >350 cells/mm3 (OR 3.9 vs. <100 cells/mm3; 95% CI 2.0–4.1, p < 0.01), total lymphocyte count >2000 cells/mm3 (OR 1.7 vs. <1000 cells/mm3; 95% CI 1.3–2.3, p < 0.01), and no prior AIDS-defining illness (OR 1.8; 95% CI 1.5–2.3, p < 0.01). Receiver-operator characteristic (ROC) analysis yielded area under the curve of 0.70 (95% CI 0.67–0.72) among derivation patients and 0.69 (95% CI 0.65–0.74) among validation patients. A cut off score >25 yielded the sensitivity of 46.7%, specificity of 79.1%, positive predictive value of 67.7%, and negative predictive value of 61.2% for prediction of pre-treatment HIV RNA <100,000 copies/mL among derivation patients.
Conclusion
A model prediction for pre-treatment HIV RNA <100,000 copies/mL produced an area under the ROC curve of 0.70. A larger sample size for prediction model development as well as for model validation is warranted.
Similar content being viewed by others
Background
Antiretroviral therapy (ART) for the treatment of human immunodeficiency virus (HIV) infection has dramatically reduced HIV-associated morbidity and mortality and has transformed HIV infection into a manageable chronic condition [1, 2]. Furthermore, early ART is highly effective in preventing HIV transmission to sexual partners [3]. More than 25 antiretroviral drugs (ARV) in 6 classes are approved for treatment of HIV infection [4]. Selection of an ARV regimen should be individualized on the basis of efficacy, adverse effects, pill burden, dosing frequency, drug–drug interactions, comorbid conditions, and cost [4, 5].
The initial ARV regimen for a treatment-naïve HIV-infected patient generally consists of 2 nucleoside/nucleotide reverse transcriptase inhibitors, usually abacavir (ABC) plus lamivudine (3TC) or tenofovir disoproxil fumarate plus emtricitabine (TDF/FTC), plus a drug from 1 of 3 drug classes: an integrase strand transfer inhibitor, a non-nucleoside reverse transcriptase inhibitor (NNRTIs), or a boosted protease inhibitor [4, 5]. ABC is usually preferred over TDF for individuals with chronic kidney disease and/or those at risk of osteoporosis and fractures [4, 5]. However, ABC is recommended for patients who are HLA-B*5701 allele negative and have a pre-treatment HIV RNA <100,000 copies/mL [6], except when used with dolutegravir (DTG) and 3TC in the same regimen [4, 5].
Rilpivirine (RPV) is a recently approved NNRTI available at relatively low cost in Thailand (7 USD per month) and other countries. The advantages of RPV are once-daily dosing and very small pill size. In addition, RPV is associated with fewer treatment discontinuations for central nervous system adverse effects, fewer lipid effects, and fewer rashes when compared with efavirenz (EFV) [7, 8]. Nevertheless, RPV has a higher rate of virological failure when compared to EFV, especially in the first 48 weeks of treatment [7]. RPV is thus recommended as an alternative option for treatment naïve HIV-infected patients with a pre-treatment HIV RNA <100,000 copies/mL and CD4 count >200 cells/mm3 [4, 5].
Testing of HIV RNA levels is recommended during initial patient visits by treatment guidelines in developed countries [4, 5]. In resource-limited settings, pre-treatment HIV RNA is not routinely performed and not widely available [9, 10]. This limits the use of ABC and RPV as a component of the first-line ARV regimen. If a clinical prediction tool based on routinely collected data could accurately predict whether pre-treatment HIV RNA was <100,000 copies/mL, this could be applied into clinical practice. The aims of this study are to determine factors associated with pre-treatment HIV RNA <100,000 copies/mL and to construct prediction tools that predict a pre-treatment HIV RNA <100,000 copies/mL. This prediction tool might support the use of ABC and RPV as part of first-line regimens for selected treatment-naïve HIV-infected individuals in resource-limited settings with limited access to HIV RNA testing.
Patients and methods
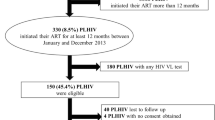
Our study population consisted of HIV-infected patients enrolled in the TREAT (Therapeutics Research, Education, and AIDS Training) Asia HIV Observational Database (TAHOD). The characteristics of this cohort have been described previously. Briefly, TAHOD is a prospective multi-center, observational study of patients with HIV and aims to assess HIV disease natural history in treated and untreated patients in the Asia and Pacific region [11]. We included patients enrolled in the cohort from 23 clinical sites throughout 13 countries in the Asia Pacific region since September 2003. The date of data censoring for the analysis of this study was 31 March 2015.
HIV-infected adults enrolled in TAHOD were eligible if they had an HIV RNA measurement documented at or around the time of ART initiation (pre-treatment HIV RNA). The window period of pre-treatment HIV RNA measurement was between 3 months prior to 1 day after the date of starting ART. ART was defined as a regimen containing ≥3 ARVs. Those exposed to mono or dual therapy prior to starting combination ART were excluded. Baseline was defined as the date of ART initiation. At baseline, co-variables included age, sex, HIV exposure, hepatitis B and C serology (ever positive), time since diagnosis of HIV infection, HIV subtype, and AIDS diagnosis prior to baseline. The window period of the following co-variables was between 3 months prior to 3 months after the date of ART initiation; body mass index (BMI), anemia (hemoglobin <13 g/dL for men, <12 g/dL for women), total lymphocyte count, CD4 count, CD8 count, CD4:CD8 ratio, and syphilis serology [Rapid plasma reagin (RPR), Venereal Disease Research Laboratory (VDRL) or Treponema pallidum particle agglutination assay (TPHA)].
Statistical analysis
The dataset was randomly split into a derivation data set (containing data from 75% of all eligible patients) and validation data set (containing data from 25% of all eligible patients) using the PROC SURVEYSELECT command in SAS version 9.4 (SAS Institute Inc., Cary, North Carolina, USA). The study endpoint was pre-treatment HIV RNA <100,000 copies/mL. Factors associated with this endpoint were evaluated by logistic regression adjusted for study site. Co-variables were considered for inclusion in the multivariate model if one or more categories exhibited a p-value <0.1. They were retained in the multivariate model if one or more categories exhibited a p-value <0.05. Missing categories, where present, were included in all models but odds ratios (OR) were not shown.
Prediction scores were created by multiplying the OR for each multivariate co-variable category by 10 and subtracting 1 [12]. Scores were rounded to the nearest 0.5 points. Some categories among the variables including in the multivariate model gave similar OR and were therefore collapsed together for the prediction tool.
The discrimination was evaluated using the area under the receiver-operator characteristic (AUROC) curve [13]. We used data of patients that had data available on all variables including in the prediction model. The optimum cut-off point for the score was evaluated by sensitivity, specificity, positive predictive value, and negative predictive value. Stata version 14.1 (StataCorp, College Station, Texas, USA) was used for all statistical analysis.
Results
A total of 2592 patients were included in our derivation analysis. Median [interquartile range (IQR)] age was 35.8 (29.9–42.5) years, 56.2% had heterosexual HIV exposure, median (IQR) BMI was 21.1 (19.0–23.4) kg/m2, median duration of HIV diagnosis was 4.3 (1.4–29.2) months, and 34.5% had prior AIDS-defining illness. Median CD4 count was 147 (50–248) cells/mm3 and median pre-treatment HIV RNA was 100,000 (34,045–301,075) copies/mL. For other laboratory investigations, 49.3% had anemia, 10.8% had positive HBsAg, 8.3% had positive anti-HCV, 19.6% had positive syphilis serology, and 75.1% had HIV infection with CRF01_AE subtype. Baseline characteristics of the patients are shown in Table 1.
Factors that statistically significantly associated with pre-treatment HIV RNA <100,000 copies/mL in the derivation patients by multivariate logistic regression, were age <30 years [OR 1.40 vs. 41–50 years; 95% confidence interval (CI) 1.10–1.80, p = 0.01], body mass index >30 kg/m2 (OR 2.4 vs. <18.5 kg/m2; 95% CI 1.1–5.1, p = 0.02), anemia (OR 1.70; 95% CI 1.40–2.10, p < 0.01], CD4 count >350 cells/mm3 (OR 3.9 vs. <100 cells/mm3; 95% CI 2.0–4.1, p < 0.01), total lymphocyte count >2000 cells/mm3 (OR 1.7 vs. <1000 cells/mm3; 95% CI 1.3–2.3, p < 0.01), and no prior AIDS-defining illness (OR 1.8; 95% CI 1.5–2.3, p < 0.01) (Table 2).
Clinical prediction tool scores for pre-treatment HIV RNA <100,000 copies/mL are shown in Table 3. Scores were +3.5 for age <30 years, +2.5 for BMI of 18.5–29.9 kg/m2 or +14.5 for BMI of >30 kg/m2, +7.0 for non-anemia, +17.0 for CD4 count >200 cells/mm3 or +5.5 for 100–199 cells/mm3, and +8.5 for no prior AIDS-defining illness. The possible maximum score was 50.5.
AUROC analysis was 0.70 (95% CI 0.67–0.72) among the derivation patients (Fig. 1) and 0.69 (95% CI 0.65–0.74) among validation patients.
A cut off total score >25 yielded sensitivity of 46.7 and 47.4%, specificity of 79.1 and 77.1%, positive predictive value of 67.7 and 64.2%, and negative predictive value of 61.2 and 63.0% for pre-treatment HIV RNA <100,000 copies/mL among the derivation patients and validation patients, respectively (Tables 4, 5). In contrast a cut off score >5 yielded the highest sensitivity of 91.1 and 91.9% and lowest specificity of 24.8 and 24.1% among derivation patients and validation patients, respectively (Tables 4, 5). We also conducted a sensitivity analysis using other prediction models, e.g. using total lymphocyte count instead of CD4 count and restriction analysis only among patients with CD4 count >200 cells/mm3, however these models did not perform better.
Discussion
Plasma HIV RNA is one laboratory test used to stage HIV disease and to assist in the selection of ARV drug regimens [4, 5]. If treatment-naïve HIV-infected patients have a pre-treatment HIV RNA >100,000 copies/mL, the following regimens are not recommended; ABC/3TC with EFV or atazanavir/ritonavir (ATV/r) or raltegravir (RAL), RPV-based regimens, and darunavir/r (DRV/r) plus RAL [4, 5]. The main reason is being the higher rates of virologic failure observed in patients who received these particular drugs [7]. In addition, patients with pre-treatment HIV RNA >100,000 copies/mL or CD4 count <200 cells/μL are a subset of patients who may experience suboptimal virologic suppression if the regimen consists of ABC or PRV [5].
To our knowledge, this is the first study on prediction tool of pre-treatment HIV RNA <100,000 copies/mL in treatment-naïve HIV-infected patients that aims to facilitate the use of ABC and RPV as one of ARV in the first-line ART in resource-limited settings. We found some clinical and laboratory factors statistically significantly associated with pre-treatment HIV RNA <100,000 copies/mL. Our prediction tool of pre-treatment HIV RNA <100,000 copies/mL performed AUROC curve of 0.70. A cut off score >25 yielded the highest specificity of 79.0% for predicting pre-treatment HIV RNA <100,000 copies/mL.
Few studies focus on the association between HIV RNA levels and HIV-related outcomes. The results from some previous studies showed that HIV RNA level is rarely directly associated with the type of opportunistic infection [14] or HIV disease progression [15]. One study demonstrated a significant correlation between HIV RNA level and wasting syndrome in naïve HIV-infected patients, with HIV RNA levels in patients with wasting syndrome, significantly higher than those without the condition [16].
We also found six independent factors associated with pre-treatment HIV RNA <100,000 copies/mL: age, BMI, anemia, CD4 count, total lymphocyte count, and prior AIDS-defining illness. For example, patients with age <30 years had higher odds of 1.4 of having pre-treatment HIV RNA <100,000 copies/mL compared to patients 41–50 years old. Furthermore, patients with baseline CD4 count 100–199 cells/mm3 had higher odds of 1.6 of having pre-treatment HIV RNA <100,000 copies/mL compared to patients with baseline CD4 count <100 cells/mm3. These factors might be easily applied in the assessment of patients in resource-limited settings because they are patients’ clinical characteristics and routine baseline laboratory investigations.
The AUROC curve is a single index for measuring the performance a test and can be used to estimate the discriminating power of a test. The AUROC of a ‘perfect’ test would be 1.00, that of a useless test, 0.50 [13, 17]. The AUROC for the pre-treatment HIV RNA model applied to the derivation population was 0.70. The AUROC curve when the model was applied to the validation population was 0.69, indicating some loss of discriminating power when applied to the new population. The score >5 showed the highest sensitivity but lowest specificity. With prediction of pre-treatment HIV RNA <100,000 copies/mL, higher specificity is required to minimize false positive results. Using a score >25 for prediction of pre-treatment HIV RNA yielded specificity approximately 80% and positive predictive value almost 70% and might be more appropriate. Additional data variables and/or an increased number of the patients might be needed to improve this prediction model and enhance its performance.
This study had some limitations. First, some patients must be excluded from the regression analysis and from the prediction tool due to missing data. Second, the performance of the model described by the AUROC of 0.70 might be associated with the small sample size of the study population among derivation and validation group.
In conclusion, in situations where HIV RNA cannot be obtained prior to ART initiation due to high costs or limited availability, certain risk factors and models for predicting pre-treatment HIV RNA <100,000 copies/mL might be useful to predict pre-treatment HIV RNA and afford opportunities for ABC and RPV initiation among naïve HIV-infected patients. A larger sample size with greater data variety would be warranted for prediction model construction as well as for model validation. Pre-treatment HIV RNA should be performed before ABC and RPV initiation if it is available and affordable.
Abbreviations
- ART:
-
antiretroviral therapy
- HIV:
-
human immunodeficiency virus
- ARV:
-
antiretroviral drugs
- ABC:
-
abacavir
- 3TC:
-
lamivudine
- TDF:
-
tenofovir disoproxil fumarate
- FTC:
-
emtricitabine
- NNRTI:
-
non-nucleoside reverse transcriptase inhibitors
- DTG:
-
dolutegravir
- RPV:
-
rilpivirine
- EFV:
-
efavirenz
- TREAT:
-
Therapeutics Research, Education, and AIDS Training
- TAHOD:
-
TREAT Asia HIV Observational Database
- BMI:
-
body mass index
- RPR:
-
rapid plasma regain
- VDRL:
-
Venereal Disease Research Laboratory
- TPHA:
-
Treponema pallidum particle agglutination assay
- OR:
-
odds ratio
- AUROC:
-
area under the receiver-operator characteristic
- IQR:
-
interquartile range
- CI:
-
confidence interval
- ATV/r:
-
atazanavir/ritonavir
- RAL:
-
raltegravir
- DRV/r:
-
darunavir/r
- CPT:
-
clinical prediction tool
- PPV:
-
positive predictive value
- NPV:
-
negative predictive value
References
Taddei TH, Lo Re V 3rd, Justice AC. HIV, aging, and viral coinfections: taking the long view. Curr HIV/AIDS Rep. 2016;13:269–78.
Kiertiburanakul S, Luengroongroj P, Sungkanuparph S. Clinical characteristics of HIV-infected patients who survive after the diagnosis of HIV infection for more than 10 years in a resource-limited setting. J Int Assoc Physicians AIDS Care (Chic). 2012;11:361–5.
Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med. 2016;375:830–9.
Panel on antiretroviral guidelines for adults and adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 14 Sep 2016.
Günthard HF, Saag MS, Benson CA, del Rio C, Eron JJ, Gallant JE, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2016 recommendations of the International Antiviral Society—USA panel. JAMA. 2016;316:191–210.
Sax PE, Tierney C, Collier AC, Fischl MA, Mollan K, Peeples L, et al. Abacavir–lamivudine versus tenofovir–emtricitabine for initial HIV-1 therapy. N Engl J Med. 2009;361:2230–40.
Cohen CJ, Molina JM, Cassetti I, Chetchotisakd P, Lazzarin A, Orkin C, et al. Week 96 efficacy and safety of rilpivirine in treatment-naive, HIV-1 patients in two phase III randomized trials. AIDS. 2013;27:939–50.
Tebas P, Sension M, Arribas J, Duiculescu D, Florence E, Hung CC, et al. Lipid levels and changes in body fat distribution in treatment-naive, HIV-1-Infected adults treated with rilpivirine or efavirenz for 96 weeks in the ECHO and THRIVE trials. Clin Infect Dis. 2014;59:425–34.
Manosuthi W, Ongwandee S, Bhakeecheep S, Leechawengwongs M, Ruxrungtham K, Phanuphak P, et al. Guidelines for antiretroviral therapy in HIV-1 infected adults and adolescents 2014 Thailand. AIDS Res Ther. 2015;12:12.
World Health Organization. Consolidated guidelines on HIV prevention, diagnosis, treatment and care for key populations. 2016. http://apps.who.int/iris/bitstream/10665/128048/1/9789241507431_eng.pdf?ua=1&ua=1. Accessed 8 Jan 2017.
Zhou J, Kumarasamy N, Ditangco R, Kamarulzaman A, Lee CK, Li PC, et al. The TREAT Asia HIV Observational Database: baseline and retrospective data. J Acquir Immune Defic Syndr. 2005;38:174–9.
Moons KG, Harrell FE, Steyerberg EW. Should scoring rules be based on odds ratios or regression coefficients? J Clin Epidemiol. 2002;55:1054–5.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36.
Kimmel AD, Goldie SJ, Walensky RP, Losina E, Weinstein MC, Paltiel AD, et al. Optimal frequency of CD4 cell count and HIV RNA monitoring prior to initiation of antiretroviral therapy in HIV-infected patients. Antivir Ther. 2005;10:41–52.
Rodríguez B, Sethi AK, Cheruvu VK, Mackay W, Bosch RJ, Kitahata M, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA. 2006;296:1498–506.
Utama MS, Merati TP. Association of opportunistic infections with HIV-RNA and CD4 cell count in pre ARV and ARV failure at the care support treatment clinic of Sanglah Hospital, Bali. J Epidemiol Res. 2016;2:13–7.
Kummar R, Indrayan A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr. 2011;48:277–89.
Authors’ contributions
SK and SS initiated concept ideas. SK, OTN, NVK, TPM, AA, WWW, MPL, RC, AK, PK, FZ, JYC, NK, RD, DDC, SO, BS, WR, PSL, EY and SP contributed data for the analysis. DB performed the statistical analysis. All authors commented on the draft manuscript. All authors read and approved the final manuscript.
Acknowledgements
PS Ly* and V Khol, National Center for HIV/AIDS, Dermatology & STDs, Phnom Penh, Cambodia; FJ Zhang*, HX Zhao and N Han, Beijing Ditan Hospital, Capital Medical University, Beijing, China; MP Lee* †, PCK Li, W Lam and YT Chan, Queen Elizabeth Hospital, Hong Kong, China; N Kumarasamy*, S Saghayam and C Ezhilarasi, Chennai Antiviral Research and Treatment Clinical Research Site (CART CRS), YRGCARE Medical Centre, VHS, Chennai, India; S Pujari*, K Joshi, S Gaikwad and A Chitalikar, Institute of Infectious Diseases, Pune, India; TP Merati*, DN Wirawan and F Yuliana, Faculty of Medicine Udayana University & Sanglah Hospital, Bali, Indonesia; E Yunihastuti*, D Imran and A Widhani, Faculty of Medicine Universitas Indonesia - Dr. Cipto Mangunkusumo General Hospital, Jakarta, Indonesia; S Oka*, J Tanuma and T Nishijima, National Center for Global Health and Medicine, Tokyo, Japan; JY Choi*, Na S and JM Kim, Division of Infectious Diseases, Department of Internal Medicine, Yonsei University College of Medicine, Seoul, South Korea; BLH Sim*, YM Gani, and R David, Hospital Sungai Buloh, Sungai Buloh, Malaysia; A Kamarulzaman*, SF Syed Omar, S Ponnampalavanar and I Azwa, University Malaya Medical Centre, Kuala Lumpur, Malaysia; R Ditangco*, E Uy and R Bantique, Research Institute for Tropical Medicine, Manila, Philippines; WW Wong*‡, WW Ku and PC Wu, Taipei Veterans General Hospital, Taipei, Taiwan; OT Ng*, PL Lim, LS Lee and PS Ohnmar, Tan Tock Seng Hospital, Singapore; A Avihingsanon*, S Gatechompol, P Phanuphak and C Phadungphon, HIV-NAT/Thai Red Cross AIDS Research Centre, Bangkok, Thailand; S Kiertiburanakul*, S Sungkanuparph, L Chumla and N Sanmeema, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand; R Chaiwarith*, T Sirisanthana, W Kotarathititum and J Praparattanapan, Research Institute for Health Sciences, Chiang Mai, Thailand; P Kantipong* and P Kambua, Chiangrai Prachanukroh Hospital, Chiang Rai, Thailand; W Ratanasuwan* and R Sriondee, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand; KV Nguyen*, HV Bui, DTH Nguyen and DT Nguyen, National Hospital for Tropical Diseases, Hanoi, Vietnam; DD Cuong*, NV An and NT Luan, Bach Mai Hospital, Hanoi, Vietnam; AH Sohn*, JL Ross* and B Petersen, TREAT Asia, amfAR—The Foundation for AIDS Research, Bangkok, Thailand; DA Cooper, MG Law*, A Jiamsakul* and DC Boettiger, The Kirby Institute, UNSW Australia, Sydney, Australia. *TAHOD Steering Committee member;†Steering Committee Chair;‡co-Chair.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available in the absence of local ethics committee approvals for the disclosure of study data, but are available from the corresponding author on reasonable request and pending study steering committee approval. Interested parties who would like to access the data underlying the findings of the study analysis may also contact research@treatasia.org for further assistance.
Ethics approval and consent to participate
Ethics approvals were obtained from institutional review boards at each of the participating clinical sites where study patient enrolment took place, as well as by separate review boards for the coordinating center (TREAT Asia, Bangkok) and the data management and analysis center (The Kirby Institute, University of New South Wales, Sydney). All patients have their data stored in both the site-level and centralized study databases for the purposes of research.
Funding
The TREAT Asia HIV Observational Database is an initiative of TREAT Asia, a program of amfAR, The Foundation for AIDS Research, with support from the US National Institutes of Health’s National Institute of Allergy and Infectious Diseases, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Cancer Institute, the National Institute of Mental Health, and the National Institute on Drug Abuse, as part of the International Epidemiologic Databases to Evaluate AIDS (IeDEA; U01AI069907). The Kirby Institute is funded by the Australian Government Department of Health and Ageing, and is affiliated with the Faculty of Medicine, UNSW Australia. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of any of the governments or institutions mentioned above.
Presentation
Parts of this manuscript were presented at the 27th European Congress of Clinical Microbiology and Infectious Diseases (Vienna), May 22–25, 2017 [abstract 784].
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Consortia
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kiertiburanakul, S., Boettiger, D., Ng, O.T. et al. Factors associated with pre-treatment HIV RNA: application for the use of abacavir and rilpivirine as the first-line regimen for HIV-infected patients in resource-limited settings. AIDS Res Ther 14, 27 (2017). https://doi.org/10.1186/s12981-017-0151-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12981-017-0151-1