Abstract
Background
Simian immunodeficiency viruses (SIVs) of chimpanzees and gorillas from Central Africa crossed the species barrier at least four times giving rise to human immunodeficiency virus type 1 (HIV-1) groups M, N, O and P. The paradigm of non-pathogenic lentiviral infections has been challenged by observations of naturally infected chimpanzees with SIVcpz associated with a negative impact on their life span and reproduction, CD4+ T-lymphocyte loss and lymphoid tissue destruction. With the advent and dissemination of new generation sequencing technologies, novel promising markers of immune deficiency have been explored in human and nonhuman primate species, showing changes in the microbiome (dysbiosis) that might be associated with pathogenic conditions. The aim of the present study was to identify and compare enteric viromes of SIVgor-infected and uninfected gorillas using noninvasive sampling and ultradeep sequencing, and to assess the association of virome composition with potential SIVgor pathogenesis in their natural hosts.
Results
We analyzed both RNA and DNA virus libraries of 23 fecal samples from 11 SIVgor-infected (two samples from one animal) and 11 uninfected western lowland gorillas from Campo-Ma’an National Park (CP), in southwestern Cameroon. Three bacteriophage families (Siphoviridae, Myoviridae and Podoviridae) represented 67.5 and 68% of the total annotated reads in SIVgor-infected and uninfected individuals, respectively. Conversely, mammalian viral families, such as Herpesviridae and Reoviridae, previously associated with gut- and several mammalian diseases were significantly more abundant (p < 0.003) in the SIVgor-infected group. In the present study, we analyzed, for the first time, the enteric virome of gorillas and their association with SIVgor status. This also provided the first evidence of association of specific mammalian viral families and SIVgor in a putative dysbiosis context.
Conclusions
Our results suggested that viromes might be potentially used as markers of lentiviral disease progression in wild gorilla populations. The diverse mammalian viral families, herein described in SIVgor-infected gorillas, may play a pivotal role in a disease progression still unclear in these animals but already well characterized in pathogenic lentiviral infections in other organisms. Larger sample sets should be further explored to reduce intrinsic sampling variation.
Similar content being viewed by others
Background
The human immunodeficiency virus (HIV) types 1 and 2 have arisen from multiple zoonotic transmissions of simian immunodeficiency viruses (SIVs) circulating in African non-human primates (NHP) to humans. SIVs from chimpanzees and gorillas from Central Africa crossed the species barrier at least four times giving rise to HIV-1 groups M, N, O, and P [1, 2]. Likewise, at least nine independent transmissions between sooty mangabeys from West Africa and humans have already been described, originating the different HIV-2 groups found in humans [3,4,5].
Although SIVs have been referred to as immunodeficiency viruses, the clinical manifestations in SIV-infected hosts were not reported during the first decades when natural infections were initially described in the wild and in naturally infected animals, captive sooty mangabeys and African green monkeys kept in zoos or primate centers. SIV lineages have already been documented in more than 40 species of African NHP [6]. Despite high SIV viral loads, individuals of these species have been initially reported as effective controllers of disease progression [7,8,9]. However, following the experimental or accidental SIV infections of Asian macaques (that are not natural reservoirs of lentiviruses), these latter primates developed an AIDS-like disease that was very similar to the human condition [10, 11].
The paradigm of non-pathogenic lentiviral infections has been recently challenged by observations of wild chimpanzees (P. t. schweinfurthii) in Gombe, Tanzania, in which infection by their natural SIV strain (SIVcpzPts) was associated with a negative impact on their life span and reproduction [12, 13]. Moreover, retrospective analysis of conserved tissues from dead SIVcpzPts-infected animals showed signs of immune deficiency similar to those found in AIDS-affected humans [13]. In addition, SIVcpz infection has also been associated with CD4+ T lymphocyte loss and lymphoid tissue destruction (alike HIV infection), leading to premature death in a naturally infected chimpanzee in a sanctuary in Cameroon [14]. Altogether, these findings underscored the need for further studies of wild NHP populations, particularly among apes, because of their highly endangered status and the potential impact of SIV infection on their survival and eventual population decline [1, 7, 15]. However, the study of these wild populations is particularly difficult and can only be carried out by noninvasive sampling.
With the advent and dissemination of next generation sequencing (NGS) technologies, novel and promising markers of immune deficiency have been explored in humans and NHP species, notably changes in the microbiome (dysbiosis) potentially associated with pathogenic conditions. In humans, HIV infection is markedly associated with expansion of enteric adenoviruses and significant loss of enteric bacterial diversity and richness, clearly involved in disease progression [16]. Similar evidence of microbiome instability was found in naturally infected chimpanzees in Gombe [17]. To present, there is no available data on the pathogenicity of SIV in gorillas. Moeller et al. [18] examining 186 fecal samples of wild gorillas from Cameroon, did not find association between SIV status and specific patterns of the gut bacteriome. Nevertheless, as gorillas are infected with SIVgor through cross-species transmission of SIVcpz from chimpanzees, it is plausible that SIVgor might negatively impact the health of gorillas in the wild [2].
By combining noninvasive sampling and NGS approaches, Handley et al. [19] reported the expansion of the enteric virome associated with disease progression in an AIDS-like context in rhesus macaques, but not with the nonpathogenic SIV infection of African green monkeys. These findings showed that pathogenic SIV infection was associated with an enteropathy that contributed to the AIDS-like disease progression seen in macaques [19]. In this report, we used a similar strategy for identifying enteric virome disturbances as dysbiosis markers in wild gorillas to understand the potential impact of SIVgor infection in their natural hosts. We identified and compared enteric viromes of SIVgor-infected and uninfected gorillas using noninvasive sampling and NGS to assess the relationship of their viromes with potential SIVgor pathogenesis. Here we present the first virome description of SIVgor-infected and uninfected gorillas, identifying viral family profiles in each studied group. Furthermore, we herein provide the first evidence of association of specific mammalian virus families (such as Adenoviridae, Herpesviridae and Reoviridae) with presence of SIVgor in a putative dysbiosis context in wild animals.
Methods
Sample collection
Twenty-three fecal samples from 22 wild western lowland gorillas (Gorilla gorilla gorilla) were studied. Samples were collected between May 2008 and February 2014 in Campo Ma’an National Park (CP), Cameroon, in a long-term follow-up project of gorilla communities in this region, where we previously reported 29% of SIVgor-infected animals in some groups [20, 21]. These samples, from 11 SIVgor-infected and 11 uninfected individuals, had been previously analyzed with serological and/or molecular tools for confirming or excluding SIVgor infection [2, 20,21,22] (Table 1). Animals were individualized with microsatellite analysis as previously described [1, 21]. SIVgor viral load (VL) was measured with a previously reported real-time RT-qPCR assay capable of quantifying all HIV-1 groups and a wide diversity of SIVcpz and SIVgor strains [23]. The fecal samples selected for the study also included being the most fresh (the shortest elapsed time from deposition to collection), having a good amount of material and also covered a wide range of VL (including the highest VL ever measured in an infected gorilla). VL estimates of SIVgor ranged from 20 to 31,497 copies per mL (cp/mL) (Table 1). Two sequential samples were collected within a time interval of approximately 1 year from a single gorilla (CPg-ID074), one sample (CP8789) in 2012, with an estimated VL of 31,497 cp/mL, and another (CP9725) in 2013, with a VL of 4924 cp/mL.
Library preparation, quantification and NGS
The NucliSens Magnetic Extraction kit (BioMerieux, Craponne, France) and QIAamp Stool DNA Miniprep Kit (QIAGEN, Courtaboeuf, France) were used to extract nucleic acids (total nucleic acids or DNA, respectively) as previously described [20, 21]. Total nucleic acids were initially treated with the TURBO DNA-free™ kit (Life Technologies, CA, USA) and purified using the RNeasy® MinElute® Cleanup kit (QIAGEN), following the manufacturers’ instructions. For removal of ribosomal RNA (from plant, bacteria and mammalian host) prior to RNA library preparation, total purified RNA was subjected to the ScriptSeq™ Complete Gold Kit: Epidemiology (Epicentre, Wisconsin, USA). The MinElute PCR Purification Kit (QIAGEN) and the Agencourt AMPure XP System: PCR Purification kit (Beckman Coulter, CA, USA) were used to purify PCR-amplified products whenever requested by the ScriptSeq™ kit protocol. DNA libraries were constructed using the Nextera® DNA Sample Preparation kit (Illumina®, CA, USA), following the manufacturer’s instructions. For all RNA and DNA library preparations, an average of 2 μg and of 50 ng of purified nucleic acid were used for each sample library, respectively. Quality control and absolute quantification of libraries were carried out in the ECO™ system (Eco Real-Time PCR System, Illumina®) using the KAPA Library Quantification kit (KAPA Biosystems, MA, USA), according to the manufacturers’ specifications. Libraries were diluted to a final concentration of 12 pM and NGS by synthesis was carried out in an Illumina® HiSeq 2500 platform (2 × 100 paired-end runs). Sequencing data files are available in the SRA database under BioProject accession PRJNA419744 (BioSample accesion: SAMN08143625—SAMN0814362547).
Bioinformatics and statistical analyses
Quality analysis of the generated sequencing reads was conducted with FastQC (Babraham Bioinformatics, Cambridge, UK) and reads with good quality (Phred score > 30 and ≥ 90 bp) were selected with Sickle-Master [25]. Reads were submitted to a BLASTX search [26] onto the GenBank nonredundant viral protein database, with a minimal e-value cutoff of 1e−5, and reads assigned to viruses were kept for further analysis. Virus taxonomic assignment was carried out using the Lowest-Common Ancestor (LCA) algorithm in MEGAN v. 6.3.5, built 4 (April 2016). The following LCA parameters were used: Min Score 50; Max Expected 1 × 10−5; Top Percent 10; Min Support Percent 75; Min Support 5; Min Complexity 0.3. Sequences without any significant hit were designated as unassigned. The MEGAN’s square-root normalization protocol enabled intra- and intergroup comparisons. We compared the virus taxa distribution between SIVgor-infected (POS) and uninfected (NEG) animals. For this and onward analyses, annotated reads of the two samples from the same SIVgor-infected animal (CPg-ID074) were pooled and averaged. Assigned sequence counts per taxa were exported for subsequent statistical analysis using GraphPad Prism v.7. Fisher’s exact test with Bonferroni correction was used for multiple comparisons. Data were presented as heatmaps with outlier data points removed whenever specified. All graphs were generated using Microsoft Excel® for Mac and R Studio v. 0.99.902. Results were considered statistically significant when p values were < 0.01.
Results
Viral diversity in enteric samples of western lowland gorillas
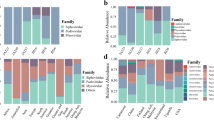
To assess the diversity of the enteric virome of wild gorillas, RNA and DNA libraries of 23 fecal samples from 11 SIVgor-infected and 11 uninfected western lowland gorillas were sequenced. We recovered 473,357,985 high-quality reads (mean = 20,580,781.96/sample, ranging from 4386,718 to 94,513,993) for all RNA libraries and 559,920,656 reads (mean = 24,344,376.35/sample, ranging from 2511,994 to 125,855,508) for all DNA libraries. After applying MEGAN square-root normalization, a reduction in read counts to 132,138.5 reads (mean = 6006.3/sample, ranging from 2575 to 14,064) was achieved for RNA libraries and to 150,539 reads (mean = 6842.7/sample, ranging from 2409 to 23,131) for DNA libraries. BLASTX analysis showed that approximately 24 and 29% of normalized reads could be annotated for identifying virus families using a viral database for RNA and DNA libraries, respectively (Fig. 1).
The Siphoviridae bacteriophage family was more frequent in SIVgor-infected individuals (32.7 vs 23.4% in uninfected individuals), while two other bacteriophage families were more frequent in uninfected individuals, Myoviridae (31.7 vs 25.8% in infected individuals) and Podoviridae (12.9 vs 9.0% in infected individuals). Altogether, these three viral families accounted for 67.5 and 68% of the total annotated reads in SIVgor-infected and uninfected individuals, respectively (Table 2). The majority of the other viral families showed frequencies below 1% of total reads. Viral family profiles diverged significantly between the two samples collected from a single individual (CPg-ID074; data not shown), a finding that precluded further analysis.
Association of gorilla virome profiles with SIVgor status
We compared the virome of the SIVgor-infected and uninfected gorillas and investigated the potential association of SIV status with virome profiles. As some viral families were represented in only one or two individuals of each group, outlier data were excluded from analysis to avoid spurious associations. In the SIVgor-infected group, 22 viral families were identified, with three taxa (Alloherpesviridae, Polydnaviridae and Reoviridae) exclusively represented in this group. Conversely, 21 viral families were identified among the uninfected gorillas, with two exclusive taxa (Microviridae and Tymoviridae). Despite the restrictive criteria of excluding outliers, we found a distinct profile for the SIVgor-infected group, with two significantly more abundant mammalian viral families (Herpesviridae and Reoviridae; p < 0.003; Table 3). The uninfected group also showed a distinct virome profile, with one significantly more abundant mammalian viral family (Rhabdoviridae; p < 0.003; Table 3). Unsupervised clustering analysis with the virome profiles of all 22 individuals revealed that some viral families served as putative proxies for SIVgor infection status, like Reoviridae and Alloherpesviridae in the SIVgor-infected group, and Tymoviridae, Microviridae and Rhabdoviridae in the uninfected group (Fig. 2).
Unsupervised clustering analysis of viral diversity profiles of SIVgor-infected (POS) and uninfected (NEG) groups, showing proxies for SIVgor infection like Reoviridae and Alloherpesviridae, associated with the POS group. Tymoviridae, Microviridae and Rhabdoviridae were associated with the NEG group
Detection of disease-associated viruses in SIVgor-infected gorillas
A specific assessment of mammalian viral families previously associated with intestinal disease in their hosts was conducted in the studied specimens. Adenoviridae reads were found only in two SIVgor-infected gorillas. The association of this specific family with prolonged infections was characterized in CPg-ID004 (VL of 655 cp/mL) and CPg-ID047 (VL of 980 cp/mL) individuals, known to be SIVgor-infected since 2007 and 2011, respectively [20, 21]. Mammalian viral families that have been associated with gut and other diseases in humans [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44] were also significantly more abundant in the SIVgor-infected group (p < 0.003), like Herpesviridae and Reoviridae, while only the Rhabdoviridae was more abundantly recovered in the uninfected group (p < 0.003).
Discussion
Studies on natural history of lentiviral infections in apes have been mostly limited to captive or semi-free ape communities due to the restrictions of working in the wild and to the endangered status of these species. However, noninvasive techniques developed during the last 30 years allowed for the study of prevalence, diversity and impact of SIV infection in several wild primate populations [6]. These advances prompted us to assess novel dysbiosis markers in previously studied wild gorilla communities, using fecal samples collected in CP.
In this study, 24 and 29% of normalized reads from RNA and DNA libraries, respectively, were annotated using a virus database. These rates were in agreement with similar reports on the complexity of virome annotation [31] and the finding of a large proportion of reads frequently found in the “No Hit” category in fecal samples [28]. These drawbacks may persist even when pretreatment for reducing bacterial or general eukaryotic material is carried out, because a substantial amount of non-viral sequences can still be found in fecal samples. It is also important to consider that a substantial proportion of the fecal virome is related to dietary components and associated viruses, like plant viruses [19, 45, 46]. In our study, a large number of annotated reads was related to non-mammalian viruses, mostly bacteriophages and plant viruses. Currently, the convergence of bacterial communities of sympatric individuals living off the same diet has been well established [19, 45, 47]. Moreover, the bacteriophage community, intrinsically linked with the bacteriome, can be an indicator of the general stability of the gut microbiome in a disease context. Mammalian viruses, on the other hand, might be important markers of disease progression despite their low frequency found during annotation.
In this study, we analyzed for the first time the enteric virome of gorillas in association with SIVgor status. This also provided the first evidence of association of specific mammalian viral families with SIVgor infection in a putative dysbiosis context. This was the case of SIV-infected rhesus macaques that presented unsuspected adenovirus infection associated with intestinal disease and enteric epithelial pathology, as well as enteric parvoviruses and advanced AIDS [19]. In the gorillas herein studied, adenoviruses were only recovered from SIVgor-infected individuals, while parvoviruses were differently represented in samples with higher VL (≥ 1000 copies/mL) than with lower VL (< 1000 copies/mL) (data not shown), suggesting that parvovirus infection might vary according to VL. The picobirnaviruses recovered in this study also showed to be more frequent in animals with higher VL than with lower VL, while reoviruses were significantly more abundant in SIVgor-infected animals than in their uninfected counterparts. Infection by picobirnaviruses and reoviruses has been associated with gastrointestinal complications in advanced HIV infections [29, 30, 41, 43]. Herpesviruses were also significantly more abundant in SIVgor-infected gorillas; these viruses have already been described in AIDS patients [36,37,38,39,40, 42] indicating persistent infections. These findings strongly suggest that SIVgor infection might be associated with dysbiosis, although further studies will be required for validation.
Epidemiological surveys showed that SIVgor as well as SIVcpz are less prevalent and considerably less evenly distributed among wild ape communities than the non-pathogenic SIVs in sooty mangabeys and African green monkeys [6]. These distribution disparities raise the possibility that apes may not be the natural reservoirs of these viruses, and that might be subjected to pathogenic conditions similar to those in other non-natural lentiviral hosts like humans and rhesus macaques [15, 19, 48, 49]. Currently SIVcpz, like HIV-1, is known to cause significant morbidity and mortality in infected chimpanzee communities [12, 13, 50], in association to compositional changes of the bacteriome rather than the virome, with disease progression with known (or suspected) immunodeficiency in late stages [51]. A similar study in wild gorillas failed to show association between SIVgor status and the bacteriome [52], while data on their virome has been, to present, wanting.
This study is underpinned by important limitations. In the great apes, it is well known that the gut microbiome community and abundance profiles are shaped by individual and external factors, like sex, age, time elapsed since fecal deposition, and dietary habits related with the rain or dry seasons of CP. With respect to sex, samples of SIVgor-infected and uninfected females showed larger numbers of reads of families of bacterial, amoebal, algal, invertebrate, and vertebrate viruses, both in SIVgor-infected and uninfected groups (not shown). In view of the small number of individuals within each group, this sex bias was not considered in further analyses. Moreover, our samples did not allow us to identify any age-related virome variation, since age estimation through noninvasive material is a very difficult, maybe unfeasible task.
Campo Ma’an is located in the southwest coast of Cameroon, a region with coastal equatorial climate where a heavy rain season (from August to November) is followed by a long dry season (from December to March), a short rain season (from April to May) and a short dry season (June and July). These alternate seasons might impose drastic diet changes resulting from different feeding strategies which, in turn, might be reflected in the fecal virome. Because we did not use seasonality as a factor for sample selection, both SIV-infected and uninfected groups had samples from dry and wet seasons, and such analysis was not considered in the present study, but deserves further assessment.
Despite the restrictive criteria herein adopted to minimize viral misidentification, the small number of studied individuals may produce imprecise estimates of microbiome profiles. Finally, the computational pipeline developed by us was characterized by a simple sequence of algorithms for identifying viral families without further characterization of full-length viral sequences or discovering novel viruses. On the other hand, the stringency for removing unreliable reads, despite hindering virus discovery, allowed robust description of the virome composition of SIVgor-infected and uninfected gorillas. However, higher number of samples will be necessary for reducing intrinsic sampling variation.
Conclusions
We herein describe, for the first time, the virome of wild gorillas, although these preliminary observations were carried out in a small number of individuals. This study suggests that the virome of SIVgor-infected individuals might differ from the one present in uninfected animals, in contrast to what has been reported for the bacteriome in this species [18]. The microbiome of SIVgor-infected animals must be further explored to validate present findings and to confirm whether virome dysbiosis might be consistent with the characteristic disease profile observed in SIVcpz-infected chimpanzees [17], the more classic pathogenic SIVmac infection in rhesus macaques, and HIV-1 infection in humans. The diverse mammalian viral families, herein described for the first time in SIVgor-infected gorillas, may play a pivotal role in a disease progression that still remains unclear in these animals but is already well characterized in pathogenic lentiviral infections. Our results suggest that viromes might be considered as markers of lentiviral disease progression in wild gorilla populations and should be further explored on a larger sample set.
References
Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–6.
D’arc M, Ayouba A, Esteban A, Learn GH, Boué V, Liegeois F, et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci. 2015;112:E1343–52.
Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, et al. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Taï Forest, Côte d’Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–27.
Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, Hahn BH. The origins of acquired immune deficiency syndrome viruses: where and when? Philos Trans R Soc Lond B Biol Sci. 2001;356:867–76.
Ayouba A, Akoua-Koffi C, Calvignac-Spencer S, Esteban A, Locatelli S, Li H, et al. Evidence for continuing cross-species transmission of SIVsmm to humans: characterization of a new HIV-2 lineage in rural Côte d’Ivoire. AIDS. 2013;27:2488–91.
Peeters M, D’Arc M, Delaporte E. The origin and diversity of human retroviruses. AIDS Rev. 2014;16:23–34.
Paiardini M, Pandrea I, Apetrei C, Silvestri G. Lessons learned from the natural hosts of HIV-related viruses. Annu Rev Med. 2009;60:485–95.
Silvestri G. Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS? J Med Primatol. 2005;34:243–52.
Silvestri G. Immunity in natural SIV infections. J Int Med. 2009;265:97–109.
Daniel MD, Letvin NL, King NW, Kannagi M, Sehgal PK, Hunt RD, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–4.
Kanki PJ, McLane MF, King NW, Letvin NL, Hunt RD, Sehgal P, et al. Serologic identification and characterization of a macaque T-lymphotropic retrovirus closely related to HTLV-III. Science. 1985;228:1199–201.
Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–9.
Rudicell RS, Holland Jones J, Wroblewski EE, Learn GH, Li Y, Robertson JD, et al. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 2010;6:e1001116.
Etienne L, Nerrienet E, LeBreton M, Bibila GT, Foupouapouognigni Y, Rousset D, et al. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology. 2011;8:4.
Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841.
Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–22.
Moeller AH, Shilts M, Li Y, Rudicell RS, Lonsdorf EV, Pusey AE, et al. SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe. 2013;14:340–5.
Moeller AH, Peeters M, Ayouba A, Ngole EM, Esteban A, Hahn BH, et al. Stability of the gorilla microbiome despite simian immunodeficiency virus infection. Mol Ecol. 2015;24:690–7.
Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–66.
Etienne L, Locatelli S, Ayouba A, Esteban A, Butel C, Liegeois F, et al. Noninvasive follow-up of simian immunodeficiency virus infection in wild-living nonhabituated western lowland gorillas in Cameroon. J Virol. 2012;86:9760–72.
Neel C, Etienne L, Li Y, Takehisa J, Rudicell RS, Bass IN, et al. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J Virol. 2010;84:1464–76.
Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164.
Etienne L, Eymard-Duvernay S, Aghokeng A, Butel C, Monleau M, Peeters M. Single real-time reverse transcription-PCR assay for detection and quantification of genetically diverse HIV-1, SIVcpz, and SIVgor strains. J Clin Microbiol. 2013;51:787–98.
Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol. 2009;83:1635–48.
Joshi N, Sickle FJ.: A sliding-window, adaptive, quality-based trimming tool for FastQ files. https://github.com/najoshi/sickle (2011). Accessed 15 Jan 2015.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71.
Cotten M, Oude Munnink B, Canuti M, Deijs M, Watson SJ, Kellam P, et al. Full genome virus detection in fecal samples using sensitive nucleic acid preparation, deep sequencing, and a novel iterative sequence classification algorithm. PLoS ONE. 2014;2:e93269.
Cunningham AL, Grohman GS, Harkness J, Law C, Marriott D, Tindall B, et al. Gastrointestinal viral infections in homosexual men who were symptomatic and seropositive for human immunodeficiency virus. J Infect Dis. 1988;158:386–91.
Gilger MA, Matson DO, Conner ME, Rosenblatt HM, Finegold MJ, Estes MK. Extraintestinal rotavirus infections in children with immunodeficiency. J Pediatr. 1992;120:912–7.
Handley SA. The virome: a missing component of biological interaction networks in health and disease. Genome Med. 2016;8:32.
Li SK, Leung RKK, Guo HX, Wei JF, Wang JH, Kwong KT, et al. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin Microbiol Infect. 2012;18:1126–33.
Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18.
Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–5.
Naeger DM, Martin JN, Sinclair E, Hunt PW, Bangsberg DR, Hecht F, et al. Cytomegalovirus-specific T cells persist at very high levels during long-term antiretroviral treatment of HIV disease. PLoS ONE. 2010;29(5):e8886.
Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012;10:655–66.
Springer KL, Weinberg A. Cytomegalovirus infection in the era of HAART: fewer reactivations and more immunity. J Antimicrob Chemother. 2004;54:582–6.
Williams B, Landay A, Presti RM. Microbiome alterations in HIV infection a review. Cell Microbiol. 2016;18:645–51.
Zou S, Caler L, Colombini-Hatch S, Glynn S, Srinivas P. Research on the human virome: where are we and what is next. Microbiome. 2016;24(4):32.
Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–9.
Giordano MO, Martinez LC, Rinaldi D, Gúinard S, Naretto E, Casero R, et al. Detection of picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:380–3.
González GG, Pujol FH, Liprandi F, Deibis L, Ludert JE. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J Med Virol. 1998;55:288–92.
Raini SK, Nyangao J, Kombich J, Sang C, Gikonyo J, Ongus JR, et al. Human rotavirus group a serotypes causing gastroenteritis in children less than 5 years and HIV-infected adults in Viwandani slum, Nairobi. Ethiop J Health Sci. 2015;25:39–46.
Stevens SJC, Blank BSN, Smits PHM, Meenhorst PL, Middeldorp JM. High Epstein–Barr virus (EBV) DNA loads in HIV-infected patients: correlation with antiretroviral therapy and quantitative EBV serology. AIDS. 2002;16:993–1001.
Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25.
Zhang T, Breitbart M, Lee WH, Run J-Q, Wei CL, Soh SWL, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3.
Moeller AH, Peeters M, Ndjango JB, Li Y, Hahn BH, Ochman H. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 2013;23:1715–20.
Hazenberg MD, Otto SA, van Benthem BHB, Roos MTL, Coutinho RA, Lange JMA, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–8.
Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity. 2010;32:737–42.
Terio KA, Kinsel MJ, Raphael J, Mlengeya T, Lipende I, Kirchhoff CA, et al. Pathologic lesions in chimpanzees (Pan trogylodytes schweinfurthii) from Gombe National Park, Tanzania, 2004–2010. J Zoo Wildl Med. 2011;42:597–607.
Barbian HJ, Li Y, Ramirez M, Klase Z, Lipende I, Mjungu D, et al. Destabilization of the gut microbiome marks the end-stage of simian immunodeficiency virus infection in wild chimpanzees. Am J Primatol. 2015. https://doi.org/10.1002/ajp.22515.
Moeller AH, Peeters M, Ayouba A, Ngole EM, Esteban A, Hahn BH, et al. Stability of the gorilla microbiome despite simian immunodeficiency virus infection. Mol Ecol. 2015;24:690–7.
Authors’ contributions
MD, MP and MAS conceived the study. MP and AA collected and provided the gorilla samples. MD processed the gorilla fecal samples for the NGS assays. MD, CF and AA carried out the construction and quantification of the RNA and DNA libraries, and ran the samples on the Illumina® HiSeq 2500 platform. MD and JS performed the data analyses. HNS, MP and MAS provided reagents and infrastructure for the conduction of the study. MD, AA, MP, HNS and MAS wrote the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgements
We wish to thank lab personnel form the Institut de Recherche pour le Développement—IRD (Montpellier, France) and Institut Bouisson Bertrand—IBB (France), mainly Dr. Eric Delaporte (Head of the Laboratory UMI233/INSERM1175 and Head of the IBB) for providing infrastructure for the conduction of the study and Amandine Esteban (technician of the laboratory UMI233/INSERM1175) for sample processing.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by grants from the Rio de Janeiro State Science Foundation (FAPERJ) E26/170.026/2008 and the Brazilian Research Council (CNPq) 573806/2008-0, both funded by the Brazilian National Institute of Science and Technology (INCT) for Cancer Control to H. N. S and M. A. S., the Graduate Program in Genetics (PGGEN) of Universidade Federal do Rio de Janeiro (UFRJ) Grant #1629742 to M. D., the National Institute of Health (RO1 AI 50529) and the Agence Nationale de Recherches sur le SIDA (ANRS 12125/12182/12325) to M. P. and A. A.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
D’arc, M., Furtado, C., Siqueira, J.D. et al. Assessment of the gorilla gut virome in association with natural simian immunodeficiency virus infection. Retrovirology 15, 19 (2018). https://doi.org/10.1186/s12977-018-0402-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12977-018-0402-9