Abstract
Background
Experimental autoimmune encephalomyelitis (EAE) is a model of inflammatory demyelinating diseases mediated by different types of leukocytes. How these cells communicate with each other to orchestrate autoimmune attacks is not fully understood, especially in the case of neutrophils, whose importance in EAE is newly established. The present study aimed to determine the expression pattern and role of different components of the IL-36 signaling pathway (IL-36α, IL-36β, IL-36γ, IL-36R) in EAE.
Methods
EAE was induced by either active immunization with myelin peptide, passive transfer of myelin-reactive T cells or injection of pertussis toxin to transgenic 2D2 mice. The molecules of interest were analyzed using a combination of techniques, including quantitative real-time PCR (qRT-PCR), flow cytometry, Western blotting, in situ hybridization, and immunohistochemistry. Microglial cultures were treated with recombinant IL-36γ and analyzed using DNA microarrays. Different mouse strains were subjected to clinical evaluation and flow cytometric analysis in order to compare their susceptibility to EAE.
Results
Our observations indicate that both IL-36γ and IL-36R are strongly upregulated in nervous and hematopoietic tissues in different forms of EAE. IL-36γ is specifically expressed by neutrophils, while IL-36R is expressed by different immune cells, including microglia and other myeloid cells. In culture, microglia respond to recombinant IL-36γ by expressing molecules involved in neutrophil recruitment, such as Csf3, IL-1β, and Cxcl2. However, mice deficient in either IL-36γ or IL-36R develop similar clinical and histopathological signs of EAE compared to wild-type controls.
Conclusion
This study identifies IL-36γ as a neutrophil-related cytokine that can potentially activate microglia, but that is only correlative and not contributory in EAE.
Similar content being viewed by others
Background
Experimental autoimmune encephalomyelitis (EAE) is an inflammatory demyelinating disease of the central nervous system (CNS) that can be induced in animals to model immunological processes involved in human diseases such as multiple sclerosis, neuromyelitis optica, and acute disseminated encephalomyelitis. EAE is initiated by CD4+ T helper lymphocytes of the Th1 or Th17 subset that recognize myelin peptides through their T cell receptors [1]. These cells do not act alone, but in concert with different myeloid cells, including monocyte-derived CD11c+ dendritic cells [2–5], which activate them by presenting the myelin peptides together with costimulatory signals [6–12], and monocyte-derived macrophages [2, 4, 13, 14], which execute effector functions leading to demyelination [15]. In addition, recent studies have established that neutrophils importantly contribute to EAE [16–19], although their precise role is still unclear.
To coordinate their actions, immune cells must communicate with each other through chemical messengers such as interleukins, chemokines, and growth factors. Many of these molecules have been studied in the context of EAE in the hope of identifying potential therapeutic targets. For example: GM-CSF derives from T cells and promotes dendritic cell recruitment [20, 21], IL-12 and IL-23 are secreted by dendritic cells to sustain T cell activation [22], and IL-1β is released from different myeloid cells to induce inflammatory responses [23]. However, no neutrophil-specific cytokine has been identified to date.
Following a study conducted to clarify the mechanism underlying the adjuvant effect of pertussis toxin (PTX) in EAE [24], we compared the transcriptional profiles of peritoneal leukocytes isolated from mice injected or not with PTX using DNA microarrays (unpublished data). Among the most upregulated genes was IL-36γ (also called IL1F9), a newly characterized cytokine of the IL-1 family [25]. The IL-36–IL-36R signaling pathway comprises three agonists (IL-36α, IL-36β, IL-36γ), two antagonists (IL-36RA, IL-38), one specific receptor subunit (IL-36R), and one accessory receptor subunit (IL-1RAcP) that is shared with the classical interleukin-1 receptor (IL-1R). The IL-36 proteins exhibit a typical β-trefoil structure [26] whose activity is enhanced by N-terminal truncation [27].
In the CNS, the expression of IL-36γ has never been studied, but that of IL-36R has been reported under normal conditions in non-neuronal elements such as the meninges, choroid plexus, and perivascular cells [28]. In the periphery, IL-36γ expression is constitutive in epithelial cell lines and inducible in myeloid cells (monocytes, macrophages, dendritic cells) by inflammatory mediators (e.g., IL-1β, TNF, IL-17, IL-23, TLR ligands) [29–37]. IL-36R is found on keratinocytes, splenic CD4+ T cells, and different myeloid cells [38–40].
IL-36γ promotes not only inflammation but also dendritic cell maturation and ability to generate a Th1 response. Indeed, recombinant IL-36γ induces cytokines and chemokines (e.g., IL-6, IL-8, Csf2, Csf3, Cxcl1, Ccl20) when injected into epithelial tissues, resulting in neutrophil and T cell infiltration [31, 40–42]. Furthermore, IL-36γ stimulates dendritic cells to produce inflammatory mediators (e.g., IL-6, IL-1β, IL-12p40, IL-12p35, IL-23p19, Cox-2, Cxcl1, Csf2) and cell surface receptors involved in antigen presentation (e.g., MHCII, CD80, CD83, CD86) [38–40]. IL-36γ can also induce CD4+ T cells to adopt an inflammatory IFNγ+ Th1 profile even in the absence of antigen-presenting cells, indicating that it can directly signal through IL-36R on T cells [38]. Consistently, the IL-36–IL-36R axis was found to be involved in psoriasis [29, 32, 34, 43–50], but the possibility that it also contributes to other T cell-driven autoimmune diseases such as multiple sclerosis remains to be determined.
The goal of the present study was to test the hypothesis that myeloid cells produces IL-36γ during EAE to promote inflammatory Th1/Th17 responses towards myelin. The findings presented below demonstrate that IL-36γ is specifically and strongly expressed by neutrophils in both CNS and hematopoietic compartments during certain forms of EAE and that microglia can react to IL-36γ by releasing neutrophil-related cytokines; however, IL-36γ does not influence the clinical and histopathological signs of EAE.
Methods
Animals
C57BL/6 mice were obtained from The Jackson Laboratory. Colonies of IL-36γ–/–, IL-36R–/–, and 2D2 mice (C57BL/6 background) were generated from breeders provided by the Mutant Mouse Regional Resource Center, Amgen or The Jackson Laboratory, respectively. The genotypes were confirmed by PCR using the primers listed in Additional file 1: Table S1. The experiments were performed on male mice aged 8–10 weeks under specific pathogen-free conditions with the approval of the Laval University Animal Protection Committee.
EAE induction by active immunization
Mice were subcutaneously injected into both flanks with a total of 200 μl of emulsion containing 300 μg of MOG35-55 peptide (Feldan) dissolved in saline and mixed with an equal volume of complete Freund’s adjuvant containing 500 μg of killed Mycobacterium tuberculosis H37 RA (Difco Laboratories). They were also injected intraperitoneally with 20 μg/kg of PTX (List Biological Laboratories) immediately and 2 days after immunization.
EAE induction by adoptive transfer
Mice were intraperitoneally injected with 20 × 106 encephalitogenic cells. These were isolated from abdominal lymph nodes and spleens of mice killed 8 days after active EAE induction and then cultured for 2 days in DMEM with MOG35-55 peptide (15 μg/ml), murine IL-12 (5 ng/ml, R&D Systems), murine IL-23 (20 ng/ml, R&D Systems), heat-inactivated HyClone bovine growth serum (10 %, Thermo Scientific), modified Eagle’s medium non-essential amino acids (1 %, Wisent), penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (250 ng/ml).
EAE induction in 2D2 mice
2D2 mice received two intraperitoneal injections of PTX (20 μg/kg) at a 2-day interval.
Evaluation of EAE symptoms
Mice were weighed and scored daily as follows: 0, no visual sign of disease; 0.5, partial tail paralysis; 1, complete tail paralysis; 1.5, weakness in one hindlimb; 2, weakness in both hindlimbs; 2.5, partial hindlimb paralysis; 3, complete hindlimb paralysis; 3.5, partial forelimb paralysis; 4, complete forelimb paralysis; and 5, dead or killed for humane reasons.
Cell suspension and flow cytometry
Mice were anesthetized and exsanguinated by cardiac perfusion with saline. Spinal cords were harvested, minced with razor blades in Dulbecco’s phosphate-buffered saline (DPBS, with Ca2+ and Mg2+), digested for 45 min at 37 °C in DPBS containing 0.13 U/ml Liberase TM (Roche Diagnostics) and 50 U/ml DNase I (Sigma-Aldrich), filtered through 40-μm cell strainers, and then separated from myelin debris by centrifugation in 35 % Percoll (GE Healthcare). The spleens were mashed through 40-μm cell strainers and treated with ammonium chloride solution (Stemcell Technologies) to remove residual erythrocytes. For immunostaining, the cells were incubated sequentially with rat anti-CD16/CD32 antibody (5 μg/ml, BD Biosciences, clone 2.4G2) and Fixable viability dye eFluor 506 (1:1000, eBioscience) for 5 min, with anti-IL-36R antibody (Abcam #ab171844 or R&D Systems #AF2354) for 30 min, and with combinations of the following antibodies for 30 min: rat anti-CD45-FITC (BD Biosciences, clone 30-F11), rat anti-CD11b-V450 (BD Biosciences, clone M1/70), rat anti-Ly6G-APC-Cy7 (Biolegend, clone 1A8), rat anti-CD3ε-PE (BD Biosciences, clone 145-2C11), rat anti-CD19-PerCP-Cy5.5, (BD Biosciences, clone 1D3), rat anti-CD11c-Alexa 647 (Biolegend, clone N418), and goat anti-rabbit IgG-Alexa 594 (Invitrogen, Cat No A11072). The latter antibodies were diluted at 1:200, except anti-CD45-FITC, which was diluted at 1:100. Isotype control antibodies and fluorescence-minus-one controls were used for gating. Cells were washed and resuspended in PBS before being analyzed with a FACSAria II flow cytometer (BD Biosciences). All the analyses were performed by excluding dead cells and doublets using FlowJo software (Tree Star, version 10.0.7r2).
Western blotting
Ly6G+ neutrophils, isolated by flow cytometry, were homogenized in extraction buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 % Triton X-100, 1 mM ethylenediaminetetraacetic acid, 1 mM ethylene glycol tetraacetic acid, 2 mM Na pyrophosphate, 10 mM Na β-glycerophosphate, 1 mM Na orthovanadate, 1 mM phenylmethanesulfonylfluoride, 1× protease and phosphatase inhibitor cocktail [Sigma]). The protein samples (50 μg) were resolved in a 12 % SDS-PAGE Mini-Protean Precast gel (Bio-Rad) and transferred to a polyvinylidene difluoride membrane (PerkinElmer). The membrane was blocked in PBS containing 0.1 % Tween 20 and 7 % non-fat milk, and then incubated at 4 °C overnight with an antibody against IL-36γ (1:200, Santa Cruz, sc-168163) or β-actin (1:50000, Abcam, mab150), followed by 1 h at room temperature in the appropriate secondary horseradish peroxidase-conjugated antibody (Cell Signaling Technology). The antibodies were detected using the Western Lightning Plus-ECL chemiluminescence substrate (Perkin Elmer).
Immunostaining
Immunostaining was performed as described previously [51] using the following primary antibodies: rat anti-CD3 (1:500, BD Biosciences, clone 17A2) and rat anti-Ly6G (1:5000, BD Biosciences, clone 1A8).
In situ hybridization
Spinal cord sections were analyzed for IL-36γ and IL-36R mRNAs by radioisotopic in situ hybridization as described previously [52].
Microscopy
Micrographs were taken using a Retiga EX camera on a Nikon E800 microscope. Images were adjusted for contrast, brightness, and sharpness using Photoshop 12 (Adobe Systems).
Microglial cell isolation and culture
Primary microglia were isolated from newborn mice (1 day old or less). The brains were minced with razor blades in DPBS and passed four times through a 20-G needle. After centrifugation (1500 rpm), the pellets were resuspended and incubated at 37 °C for 15 min in DPBS containing 0.25 % trypsin-EDTA (Wisent) and 50 U/ml DNase I. The cells were plated in T-25 flasks (~3 brains/flask) and grown in Dulbecco’s modified Eagle’s medium (Wisent) supplemented with 10 % heat-inactivated HyClone bovine growth serum, with medium changed every 2 days. After 2 weeks, the microglia were separated from other glial cells as described previously [53]. Briefly, they were incubated with 0.05 % trypsin in DMEM at 37 °C for 30–45 min, rinsed with PBS to remove contaminating non-adherent cells, then grown in a 1:1 mixture of fresh media and conditioned media from mixed glial cultures (2–3 days). Microglial purity was >99 %, as determined by flow cytometry using the myeloid cell marker CD11b.
For experimentation, primary microglia were seeded in 12-well plates and used at ~70 % confluence (after 5–7 days). BV2 microglia were seeded in 12-well plates at a density of 500,000 cells and used on the next day. Cells were stimulated with 100 ng/ml recombinant mouse IL-36γ (amino acids 13-164, R&D Systems) or PBS as control. After 6 h, the cells were rinsed with PBS, resuspended in lysis buffer and stored at −80 °C until RNA extraction.
RNA extraction and qRT-PCR
Total RNA from tissues and cells was isolated by homogenization in TRI-reagent or lysis buffer (Sigma-Aldrich), respectively. GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) was used for RNA purification. First strand cDNA was generated from 5 (tissues) or 1 (cells) μg of total RNA using Superscript III (Invitrogen) with random hexamers and 20-mer oligo-dT primers, then purified using the GenElute PCR Clean-Up Kit (Sigma-Aldrich). The product (20 ng) was analyzed using the LightCycler 480 system with the SYBR Green I Master mix and primers listed in Additional file 2: Table S2 according to the manufacturer’s instructions (Applied Biosystems). The PCR conditions consisted of 45 cycles of denaturation (10 s at 95 °C), annealing (10 s at 60 °C), elongation (14 s at 72 °C), and reading (5 s at 74 °C). The number of mRNA copies was determined using the second derivative method [54].
Statistical analyses
Data are expressed as mean ± standard error. Means were compared using the Wilcoxon or Kruskal-Wallis test, except for the microarray data, which were compared using Student’s t test. EAE incidence curves were constructed using the Kaplan-Meier method and compared using the Wilcoxon test. All these analyses were performed using JMP 10 (SAS Institute) with a significance level of 5 %.
Results
Neutrophils express IL-36γ in the CNS and hematopoietic tissues of EAE mice
To compare the spatio-temporal expression patterns of IL-36R and its agonists (IL-36α, IL-36β, IL-36γ) in different MS models, we quantified their mRNAs by quantitative real-time PCR (qRT-PCR) in nervous and hematopoietic tissues from mice killed at various time points after EAE induction by either one of the following methods: (1) active immunization with MOG35-55 peptide in complete Freund’s adjuvant (CFA) plus PTX (active EAE), (2) adoptive transfer of splenocytes isolated from EAE mice on day 8 and restimulated in culture for 2 days with MOG35-55 in the presence of IL-12 and IL-23 (passive EAE), and (3) intraperitoneal injections of PTX to 2D2 mice expressing a transgenic MOG-specific T cell receptor [55]. Because the latter mice develop EAE within 2 weeks at an incidence lower than 60 % [24, 55], we killed them at a single time point (day 15) and separated them into those that had developed EAE or not (2D2 with or without EAE). Wild-type mice injected with PBS, PTX, or CFA only were used as controls.
In the spinal cord, IL-36γ and IL-36R mRNAs were markedly increased in the three EAE models, compared to control mice and PTX-treated 2D2 mice without EAE (Fig. 1). An important difference between the active and passive models is that IL-36γ mRNA peaked at the latest time point examined (day 12) in the former, whereas it peaked at day 9 and declined thereafter in the latter. In the hematopoietic tissues (spleen, blood), IL-36γ mRNA was increased in active EAE and, to a lesser extent, in response to PTX and CFA, but not in passive EAE. The upregulation of IL-36γ was MOG-specific in the spleen and spinal cord, but mainly attributable to the adjuvants in the blood. Higher expression of IL-36γ mRNA was also found in the spleen of 2D2 mice compared to that of wild-type mice, but there was no difference between PTX-treated 2D2 mice with or without EAE. IL-36R mRNA was modestly increased only in the spleen of mice with active EAE. No significant expression of IL-36α and IL-36β mRNAs was detected in any of the tissues and conditions examined (average amounts detected: <220 copies per μm of total RNA; data not shown). Together, these results suggest that IL-36γ is the primary IL-36R agonist that is upregulated during EAE in both CNS and hematopoietic tissues, where its receptor can also be expressed.
IL-36γ and IL-36R are transcriptionally upregulated in 3 EAE models. Quantification of the mRNAs encoding IL-36γ and IL-36R by qRT-PCR in different tissues from mice killed at the indicated time points after induction of EAE by either active immunization with MOG peptide, passive transfer of encephalitogenic T cells, or injection of PTX to 2D2 mice. Control non-transgenic mice were injected with PBS, PTX, or CFA only. Stars indicate significant differences from the PBS group only (white star) or all the corresponding control groups (black star), as determined by Wilcoxon tests (P ≤ 0.038). Sample size: 11–13 (PBS), 5–13 (PTX), 4–8 (CFA), 7–8 (active EAE), 5–8 (passive EAE), 6 (2D2 with EAE), or 5 (2D2 without [w/o] EAE)
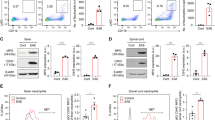
To identify the cell types expressing IL-36γ and IL-36R, we first quantified their mRNAs by qRT-PCR in different leukocytes purified from the spleen or spinal cord of mice with active EAE (day 12) by flow cytometry on the basis of established cell-specific markers. In both tissues, IL-36γ mRNA was detected at high levels only in Ly6G+ neutrophils, whereas IL-36R was detected in different cells, especially those of the myeloid lineage (Fig. 2a). Second, the expression of IL-36γ was confirmed at the protein level by Western blotting using FACS-purified splenic neutrophils (note that it was not possible to include neutrophils from spinal cords, because they were less abundant and yielded not enough proteins). A specific band for IL-36γ was detected at the predicted molecular weight of ~22 kDa in neutrophils from IL-36γ+/+ mice, but not from IL-36γ−/− mice (Fig. 2b). We also attempted to confirm the expression of IL-36R by flow cytometry using two different antibodies, but none of them provided reliable results, as the labeling was comparable between IL-36R+/+ and IL-36R−/− neutrophils and was therefore non-specific (data not shown). Third, we examined the distribution of IL-36γ mRNA in spinal cord sections from EAE and naïve mice by radioisotopic in situ hybridization. Some of these sections were also immunostained for Ly6G or the macrophage/microglia marker Iba1. Both transcripts were strongly expressed in EAE mice in areas near the meninges, but were not detectable in naïve mice (Fig. 2c). IL-36γ mRNA was co-localized with Ly6G+ cells, while IL-36R mRNA was reliably co-localized only with Iba1+ cells (Fig. 2d), although we could not exclude its presence in other cell types. These results led us to conclude that neutrophils can produce IL-36γ during EAE in both CNS and peripheral compartments, potentially to communicate with microglia and other myeloid cells.
IL-36γ is selectively expressed by neutrophils, while IL-36R is expressed by different leukocytes such as monocytic cells. a Quantification of IL-36γ or IL36R mRNA by qRT-PCR in different cells purified from the spinal cord or spleen of EAE mice by FACS using the following gating strategies: neutrophils, Ly6G+CD11b+CD45+CD3−CD19−; microglia, CD11b+CD45lowLy6G−CD3−CD19−; monocyte-derived cells (MDC; comprising macrophages and CD11c+ dendritic cells), CD11b+CD45+CD11c+/−Ly6G−CD3−CD19−; other intraspinal leukocytes, CD45+CD3+/−CD19+/−CD11b−Ly6G−CD11c−; B cells, CD19+CD45+CD11b−Ly6G−CD3−; splenic macrophages, CD11b+CD45+CD11c−Ly6G−CD3−CD19−; other splenic leukocytes, CD45+CD3+/−CD11b+/−CD11c+/−Ly6G−CD19−. Sample size: spinal cord, one pooled sample of sorted cells from four mice; spleen, five non-pooled samples from individual mice. b Western blotting showing the full-length form of IL-36γ (~22 kDa) in splenic neutrophils from a wild-type EAE mouse (IL-36γ+/+), but not from an IL-36γ-deficient EAE mouse (IL-36γ−/−). Data are representative of at least four mice per group. The recombinant (truncated) form of IL-36γ was used as a control (right lane). β-actin (lower panels) was used to control for protein loading. Asterisks indicate non-specific bands. c Autoradiograms showing in situ hybridization signals (arrows) for IL-36γ or IL-36R mRNA in the spinal cord of mice killed on day 12 after active EAE induction, but not in naïve mice. Note the submeningeal distribution of the signals (representative of at least five mice). Scale bar = 500 μm. d Double labeling for IL-36γ or IL-36R mRNA (black grains, in situ hybridization) and cell type-specific markers (red brown, immunohistochemistry) in CNS sections from EAE mice. Arrows show examples of double-labeled cells. Scale bar = 20 μm
Microglia respond to IL-36γ by expressing inflammatory genes
To demonstrate that microglia are adequately equipped to respond to IL-36γ, we first compared the transcriptional profiles of BV2 microglial cells treated or not for 6 h with recombinant IL-36γ using Affymetrix DNA microarrays. The dose used (100 ng/ml) was chosen because it was previously reported to be optimal for stimulating IL-6 secretion from cultured dendritic cells [38]. Twenty-two genes were found to be upregulated ≥2 times with a hybridization signal ≥100 (Additional file 3: Table S3 and Fig. 3a). The three most upregulated genes coded for Csf3, IL-1β, and Cxcl2, which are well known to be involved in neutrophil biology [56]. Quantitative PCR analysis confirmed that these 3 genes were strongly upregulated in BV2 cells by IL-36γ (Fig. 3b). To validate these results, we repeated the experiment with primary microglia collected from IL-36R+/+ and IL-36R−/− mice. As expected, upregulation of Csf3, IL-1β, and Cxcl2 were observed in IL-36R+/+ microglia following IL-36γ exposure, but not in IL-36R−/− microglia (Fig. 3c). The latter cells were not devoid of the ability to express these genes because they expressed them at the same levels as IL-36R+/+ microglia when stimulated with lipopolysaccharide (10 ng/ml; data not shown). Altogether, these results indicate that microglia have the potential to respond to IL-36γ, at least ex vivo.
IL-36γ stimulates the expression of inflammatory genes in microglial cultures. a Genes that were upregulated ≥2.5 times in BV2 microglia treated for 6 h with recombinant IL-36γ (100 ng/ml) compared to PBS, as determined using Affymetrix DNA microarrays (Student’s t test, P < 0.04). b, c Quantitative PCR analysis confirming that Csf3, IL-1β, and Cxcl2 mRNAs were upregulated by IL-36γ in BV2 cells (b) and primary microglia from IL-36R+/+ mice (c) (Wilcoxon test, P < 0.005). Note in c the absence of upregulation in primary microglia from IL-36R−/− mice, confirming the specificity of the results
The IL-36γ–IL-36R axis does not influence the course of active EAE
As IL-36γ and IL-36R are strongly and chronically expressed in active EAE (Figs. 1 and 2) and as the resident macrophages of the CNS can potentially respond to IL-36γ (Fig. 3), we hypothesized that the loss of these proteins would have an impact on the clinical course of active EAE. To address this possibility, we induced the disease in mice deficient or not in either IL-36γ or IL-36R and then scored their symptoms daily for 21 days. As shown in Fig. 4, the incidence, onset, and severity of EAE were similar between the knockouts and their wild-type controls. To examine whether the genetic deletions had influenced the nature of the immune cells infiltrating the CNS, we killed all of the mice on day 21 and prepared single-cell suspensions from their spinal cords for flow cytometric analysis. Consistently with the behavioral observations, no intergenotype difference was observed in the number of microglia, macrophages, dendritic cells, neutrophils as well as of T and B cells (Fig. 5). Therefore, these results demonstrate that the IL-36γ-IL-36R axis does not play a significant role in the autoimmune response involved in active EAE.
Neither IL-36γ nor IL-36R is required for EAE development. a Kaplan-Meier curves showing EAE incidence in mice expressing (black squares) or lacking (white squares) IL-36γ (upper graph) or IL-36R (bottom graph) after immunization with MOG. The graphs include all the mice tested. No significant intergenotype difference was detected (Wilcoxon tests, P ≥ 0.41). Sample size: 20 (IL-36γ+/+), 20 (IL-36γ−/−), 6 (IL-36R+/+), or 6 (IL-36R−/−). b EAE severity in mice expressing (black squares) or lacking (white squares) IL-36γ (upper graph) or IL-36R (bottom graph). The graphs include only mice that had developed clinical signs of EAE at the end of the study (i.e., 21 days). No significant intergenotype difference was detected (Wilcoxon tests, P ≥ 0.12). Sample size: 17 (IL-36γ+/+), 17 (IL-36γ−/−), 5 (IL-36R+/+), or 5 (IL-36R−/−). c Additional statistics for EAE in mice expressing (+/+) or not (−/−)IL-36γ or IL-36R. No significant intergenotype difference was detected in any of these parameters (Wilcoxon tests, P ≥ 0.34). Sample size: 20 (IL-36γ+/+), 20 (IL-36γ−/−), 6 (IL-36R+/+), or 6 (IL-36R−/−)
No difference in leukocyte recruitment in the spinal cord of EAE mice expressing or not IL-36γ or IL-36R. a Gating strategy used for flow cytometric analysis. Dead cells and doublets were excluded. The data shown are from a representative wild-type EAE mouse. b, c Counts of immune cells in the spinal cord of IL-36γ−/−, IL-36R−/−, or wild-type mice killed 21 days after immunization with MOG peptide. The counts were normalized to CD45− cells used as an internal control. When the cells were collected, the animals had clinical scores ranging from 0.5 to 3. No significant difference was found between the genotypes (Wilcoxon tests, P ≥ 0.07). Sample size: 6 (b) or 9 (c) mice per group
Discussion
As observed in other immunological diseases such as psoriasis [29, 32, 34, 57, 58], the present study shows that IL-36γ and IL-36R are upregulated in the three models of EAE. It also shows that IL-36γ derives from neutrophils and can stimulate microglia to produce neutrophil-stimulating cytokines, which is consistent with previous studies showing that IL-36γ induces both the production of such cytokines in other myeloid cells residing outside the CNS [31, 38, 40–42] and the recruitment of neutrophils [42]. Despite these observations, we demonstrate that neither IL-36γ nor IL-36R plays a significant role in EAE, similarly to what was reported in murine models of arthritis [59, 60] and mycobacterial infection [61].
The main question that arises is why IL-36γ is expressed, but does not play a role in certain conditions. One hypothesis is that IL-36γ, like other members of the IL-1 family, requires a second signal to be processed, activated, and secreted. This signal would be present in psoriasis, but not in EAE and arthritis models. In support of this, it has been shown that IL-36γ requires N-terminal truncation to be fully active [27]. However, it is unknown whether this truncation occurs in vivo and what would be the protease involved. Our work shows, for the first time to our knowledge, that the full-length IL-36γ protein (193 amino acids, 22 kDa) is detectable in vivo (i.e., in neutrophils from EAE mice). As the truncated form is not detectable in the same conditions, it is likely that IL-36γ remains stored under a pro-form in the cytoplasm of neutrophils during EAE. A second (non-exclusive) hypothesis is that neutrophils release IL-36γ during a cell death process such as NETosis, which occurs in psoriasis [62], but not likely in EAE. This possibility that IL-36γ acts as an alarmin (i.e., an endogenous molecule that signals tissue and cell damage) is so far only supported by the observation that the TLR3 agonist polyinosinic-polycytidylic acid concomitantly induces cell death and IL-36γ release in keratinocyte cultures [35]. Finally, a third hypothesis is that the CNS, an immunoprivileged site, has a higher activation threshold to IL-36γ due to a combination of mechanisms (e.g., via the secretion of IL-36 antagonists and the generation of intracellular signaling inhibitors such as SIGIRR [63]).
A positive aspect of this study is that we have identified microglia as a potential target of IL-36γ. This suggests that IL-36γ could contribute to neuroinflammation, perhaps by promoting neutrophil recruitment, in certain conditions that remain to be determined. Another positive aspect is that we have identified neutrophils as a major and exclusive source of IL-36γ in nervous and hematopoietic tissues (i.e., brain, spinal cord, spleen, blood). According to previous studies, keratinocytes and other epithelial cells can also produce IL-36γ in culture [30–34, 64], but, to our knowledge, this remains to be confirmed in vivo. Therefore, it will be important in the future to determine whether neutrophils are a predominant source of IL-36γ in epithelial inflammatory diseases (e.g., psoriasis, asthma) as they are in EAE.
In conclusion, this study demonstrates that components of the IL-36 signaling pathway are strongly expressed in EAE, but that they do not contribute to this pathology. Furthermore, by revealing that IL-36γ is a neutrophil-specific marker in different tissues and that microglia have the potential to response to this cytokine, this study opens up new research avenues for elucidating the biological function of IL-36γ and its clinical significance.
References
Goverman J. Autoimmune T cell responses in the central nervous system. Nat Rev Immunol. 2009;9:393–407.
King IL, Dickendesher TL, Segal BM. Circulating Ly-6C+ myeloid precursors migrate to the CNS and play a pathogenic role during autoimmune demyelinating disease. Blood. 2009;113:3190–7.
Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Capello E, Mancardi GL, et al. Dendritic cells in multiple sclerosis lesions: maturation stage, myelin uptake, and interaction with proliferating T cells. J Neuropathol Exp Neurol. 2006;65:124–41.
Fischer HG, Reichmann G. Brain dendritic cells and macrophages/microglia in central nervous system inflammation. J Immunol. 2001;166:2717–26.
Serafini B, Columba-Cabezas S, Di Rosa F, Aloisi F. Intracerebral recruitment and maturation of dendritic cells in the onset and progression of experimental autoimmune encephalomyelitis. Am J Pathol. 2000;157:1991–2002.
McMahon EJ, Bailey SL, Castenada CV, Waldner H, Miller SD. Epitope spreading initiates in the CNS in two mouse models of multiple sclerosis. Nat Med. 2005;11:335–9.
Greter M, Heppner FL, Lemos MP, Odermatt BM, Goebels N, Laufer T, et al. Dendritic cells permit immune invasion of the CNS in an animal model of multiple sclerosis. Nat Med. 2005;11:328–34.
Deshpande P, King IL, Segal BM. Cutting edge: CNS CD11c + cells from mice with encephalomyelitis polarize Th17 cells and support CD25 + CD4+ T cell-mediated immunosuppression, suggesting dual roles in the disease process. J Immunol. 2007;178:6695–9.
Bailey SL, Schreiner B, McMahon EJ, Miller SD. CNS myeloid DCs presenting endogenous myelin peptides ‘preferentially’ polarize CD4+ T(H)-17 cells in relapsing EAE. Nat Immunol. 2007;8:172–80.
Kivisakk P, Imitola J, Rasmussen S, Elyaman W, Zhu B, Ransohoff RM, et al. Localizing central nervous system immune surveillance: meningeal antigen-presenting cells activate T cells during experimental autoimmune encephalomyelitis. Ann Neurol. 2009;65:457–69.
Bartholomaus I, Kawakami N, Odoardi F, Schlager C, Miljkovic D, Ellwart JW, et al. Effector T cell interactions with meningeal vascular structures in nascent autoimmune CNS lesions. Nature. 2009;462:94–8.
Wlodarczyk A, Lobner M, Cedile O, Owens T. Comparison of microglia and infiltrating CD11c(+) cells as antigen presenting cells for T cell proliferation and cytokine response. J Neuroinflammation. 2014;11:57.
Tran EH, Hoekstra K, van Rooijen N, Dijkstra CD, Owens T. Immune invasion of the central nervous system parenchyma and experimental allergic encephalomyelitis, but not leukocyte extravasation from blood, are prevented in macrophage-depleted mice. J Immunol. 1998;161:3767–75.
Heppner FL, Greter M, Marino D, Falsig J, Raivich G, Hovelmeyer N, et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat Med. 2005;11:146–52.
Yamasaki R, Lu H, Butovsky O, Ohno N, Rietsch AM, Cialic R, et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med. 2014;211:1533–49.
Carlson T, Kroenke M, Rao P, Lane TE, Segal B. The Th17-ELR+ CXC chemokine pathway is essential for the development of central nervous system autoimmune disease. J Exp Med. 2008;205:811–23.
Liu L, Darnall L, Hu T, Choi K, Lane TE, Ransohoff RM. Myelin repair is accelerated by inactivating CXCR2 on nonhematopoietic cells. J Neurosci. 2010;30:9074–83.
Roy M, Richard JF, Dumas A, Vallieres L. CXCL1 can be regulated by IL-6 and promotes granulocyte adhesion to brain capillaries during bacterial toxin exposure and encephalomyelitis. J Neuroinflammation. 2012;9:18.
Aube B, Levesque SA, Pare A, Chamma E, Kebir H, Gorina R, et al. Neutrophils mediate blood-spinal cord barrier disruption in demyelinating neuroinflammatory diseases. J Immunol. 2014;193:2438–54.
Codarri L, Gyülvészi G, Tosevski V, Hesske L, Fontana A, Magnenat L, et al. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–7.
El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–75.
Kroenke MA, Carlson TJ, Andjelkovic AV, Segal BM. IL-12- and IL-23-modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. J Exp Med. 2008;205:1535–41.
Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–32.
Dumas A, Amiable N, de Rivero Vaccari JP, Chae JJ, Keane RW, Lacroix S, et al. The inflammasome pyrin contributes to pertussis toxin-induced IL-1beta synthesis, neutrophil intravascular crawling and autoimmune encephalomyelitis. PLoS Pathog. 2014;10, e1004150.
Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–18.
Gunther S, Sundberg EJ. Molecular determinants of agonist and antagonist signaling through the IL-36 receptor. J Immunol. 2014;193:921–30
Towne JE, Renshaw BR, Douangpanya J, Lipsky BP, Shen M, Gabel CA, et al. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36alpha, IL-36beta, and IL-36gamma) or antagonist (IL-36Ra) activity. J Biol Chem. 2011;286:42594–602.
Lovenberg TW, Crowe PD, Liu C, Chalmers DT, Liu XJ, Liaw C, et al. Cloning of a cDNA encoding a novel interleukin-1 receptor related protein (IL 1R-rp2). J Neuroimmunol. 1996;70:113–22.
Debets R, Timans JC, Homey B, Zurawski S, Sana TR, Lo S, et al. Two novel IL-1 family members, IL-1 delta and IL-1 epsilon, function as an antagonist and agonist of NF-kappa B activation through the orphan IL-1 receptor-related protein 2. J Immunol. 2001;167:1440–6.
Vos JB, van Sterkenburg MA, Rabe KF, Schalkwijk J, Hiemstra PS, Datson NA. Transcriptional response of bronchial epithelial cells to Pseudomonas aeruginosa: identification of early mediators of host defense. Physiol Genomics. 2005;21:324–36.
Chustz RT, Nagarkar DR, Poposki JA, Favoreto SJ, Avila PC, Schleimer RP, et al. Regulation and function of the IL-1 family cytokine IL-1F9 in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2011;45:145–53.
Johnston A, Xing X, Guzman AM, Riblett M, Loyd CM, Ward NL, et al. IL-1F5, -F6, -F8, and -F9: a novel IL-1 family signaling system that is active in psoriasis and promotes keratinocyte antimicrobial peptide expression. J Immunol. 2011;186:2613–22.
Muhr P, Zeitvogel J, Heitland I, Werfel T, Wittmann M. Expression of interleukin (IL)-1 family members upon stimulation with IL-17 differs in keratinocytes derived from patients with psoriasis and healthy donors. Br J Dermatol. 2011;165:189–93.
Carrier Y, Ma HL, Ramon HE, Napierata L, Small C, O’Toole M, et al. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: implications in psoriasis pathogenesis. J Invest Dermatol. 2011;131:2428–37.
Lian LH, Milora KA, Manupipatpong KK, Jensen LE. The double-stranded RNA analogue polyinosinic-polycytidylic acid induces keratinocyte pyroptosis and release of IL-36γ. J Invest Dermatol. 2012;132:1346–53.
Bachmann M, Scheiermann P, Hardle L, Pfeilschifter J, Muhl H. IL-36gamma/IL-1F9, an innate T-bet target in myeloid cells. J Biol Chem. 2012;287:41684–96.
Gresnigt MS, Rosler B, Jacobs CW, Becker KL, Joosten LA, van der Meer JW, et al. The IL-36 receptor pathway regulates Aspergillus fumigatus-induced Th1 and Th17 responses. Eur J Immunol. 2013;43:416–26.
Vigne S, Palmer G, Lamacchia C, Martin P, Talabot-Ayer D, Rodriguez E, et al. IL-36R ligands are potent regulators of dendritic and T cells. Blood. 2011;118:5813–23.
Mutamba S, Allison A, Mahida Y, Barrow P, Foster N. Expression of IL-1Rrp2 by human myelomonocytic cells is unique to DCs and facilitates DC maturation by IL-1F8 and IL-1F9. Eur J Immunol. 2012;42:607–17.
Foster AM, Baliwag J, Chen CS, Guzman AM, Stoll SW, Gudjonsson JE, et al. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J Immunol. 2014;192:6053–61.
Towne JE, Garka KE, Renshaw BR, Virca GD, Sims JE. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J Biol Chem. 2004;279:13677–88.
Ramadas RA, Ewart SL, Medoff BD, LeVine AM. Interleukin-1 family member 9 stimulates chemokine production and neutrophil influx in mouse lungs. Am J Respir Cell Mol Biol. 2011;44:134–45.
Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204:2603–14.
Blumberg H, Dinh H, Dean CJ, Trueblood ES, Bailey K, Shows D, et al. IL-1RL2 and its ligands contribute to the cytokine network in psoriasis. J Immunol. 2010;185:4354–62.
Onoufriadis A, Simpson MA, Pink AE, Di Meglio P, Smith CH, Pullabhatla V, et al. Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am J Hum Genet. 2011;89:432–7.
Sugiura K, Takeichi T, Kono M, Ogawa Y, Shimoyama Y, Muro Y, et al. A novel IL36RN/IL1F5 homozygous nonsense mutation, p.Arg10X, in a Japanese patient with adult-onset generalized pustular psoriasis. Br J Dermatol. 2012;167:699–701.
Tortola L, Rosenwald E, Abel B, Blumberg H, Schafer M, Coyle AJ, et al. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J Clin Invest. 2012;122:3965–76.
Sugiura K, Takemoto A, Yamaguchi M, Takahashi H, Shoda Y, Mitsuma T, et al. The majority of generalized pustular psoriasis without psoriasis vulgaris is caused by deficiency of interleukin-36 receptor antagonist. J Invest Dermatol. 2013;133:2514–21.
Farooq M, Nakai H, Fujimoto A, Fujikawa H, Matsuyama A, Kariya N, et al. Mutation analysis of the IL36RN gene in 14 Japanese patients with generalized pustular psoriasis. Hum Mutat. 2013;34:176–83.
Kanazawa N, Nakamura T, Mikita N, Furukawa F. Novel IL36RN mutation in a Japanese case of early onset generalized pustular psoriasis. J Dermatol. 2013;40:749–51.
Audoy-Remus J, Richard JF, Soulet D, Zhou H, Kubes P, Vallieres L. Rod-Shaped monocytes patrol the brain vasculature and give rise to perivascular macrophages under the influence of proinflammatory cytokines and angiopoietin-2. J Neurosci. 2008;28:10187–99.
Bedard A, Tremblay P, Chernomoretz A, Vallieres L. Identification of genes preferentially expressed by microglia and upregulated during cuprizone-induced inflammation. Glia. 2007;55:777–89.
Saura J, Tusell JM, Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–9.
Luu-The V, Paquet N, Calvo E, Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques. 2005;38:287–93.
Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–81.
Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–75.
D’Erme AM, Wilsmann-Theis D, Wagenpfeil J, Hölzel M, Ferring-Schmitt S, Sternberg S, et al. IL-36γ (IL-1F9) is a biomarker for psoriasis skin lesions. J Invest Dermatol. 2015;135:1025–32.
Keermann M, Kõks S, Reimann E, Prans E, Abram K, Kingo K. Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC Genomics. 2015;16:322.
Lamacchia C, Palmer G, Rodriguez E, Martin P, Vigne S, Seemayer CA, et al. The severity of experimental arthritis is independent of IL-36 receptor signaling. Arthritis Res Ther. 2013;15:R38.
Derer A, Groetsch B, Harre U, Böhm C, Towne J, Schett G, et al. Blockade of IL-36 receptor signaling does not prevent from TNF-induced arthritis. PLoS One. 2014;9, e101954.
Segueni N, Vigne S, Palmer G, Bourigault ML, Olleros ML, Vesin D, et al. Limited contribution of IL-36 versus IL-1 and TNF pathways in host response to mycobacterial infection. PLoS One. 2015;10, e0126058.
Pinegin B, Vorobjeva N, Pinegin V. Neutrophil extracellular traps and their role in the development of chronic inflammation and autoimmunity. Autoimmun Rev. 2015;14:633–40.
Costelloe C, Watson M, Murphy A, McQuillan K, Loscher C, Armstrong ME, et al. IL-1F5 mediates anti-inflammatory activity in the brain through induction of IL-4 following interaction with SIGIRR/TIR8. J Neurochem. 2008;105:1960–9.
Kumar S, McDonnell PC, Lehr R, Tierney L, Tzimas MN, Griswold DE, et al. Identification and initial characterization of four novel members of the interleukin-1 family. J Biol Chem. 2000;275:10308–14.
Acknowledgements
This work was supported by the Multiple Sclerosis Society of Canada and the Canadian Institutes for Health Research. LB received a Merit Scholarship from the Fonds de recherche du Québec—Nature et technologies.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LV designed the experiments and supervised the project. LB carried out most of the experiments with the assistance of AD and AP. LV and LB wrote the manuscript, which was read and approved by the other authors.
Additional files
Additional file 1: Table S1.
Primers used for genotyping. (PDF 39 kb)
Additional file 2: Table S2.
Primers used for qRT-PCR. (PDF 38 kb)
Additional file 3: Table S3.
Genes that were upregulated ≥2 times in BV2 microglial cells cultured for 6 h with recombinant IL-36γ (100 ng/ml), as determined using Affymetrix DNA microarrays. (PDF 70 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bozoyan, L., Dumas, A., Patenaude, A. et al. Interleukin-36γ is expressed by neutrophils and can activate microglia, but has no role in experimental autoimmune encephalomyelitis. J Neuroinflammation 12, 173 (2015). https://doi.org/10.1186/s12974-015-0392-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12974-015-0392-7