Abstract
Background
-
Context: Methotrexate (MTX) is a cornerstone in the treatment of juvenile idiopathic arthritis (JIA). MTX treatment is commonly associated with nausea. Large inter-individual variation exists in the level of MTX-induced nausea, possibly due to genetic factors.
-
Purpose: To investigate whether MTX-induced nausea was associated with single nucleotide polymorphisms (SNPs) in genes encoding MTX-transporter proteins, a MTX metabolizing enzyme and a nausea receptor.
Findings
-
Methods: Children aged ≥9 years treated with MTX for JIA were eligible. MTX-induced nausea was registered by the children’s completion of a nausea diary (min. 7 days) and the parents’ completion of the MTX intolerance severity score (MISS). The selected SNPs were: SLCO1B1 (rs4149056; rs4149081), SLCO1B3 (rs2117032), SLC19A1 (rs1051266), ABCC2 (rs2273697; rs3740066; rs717620), ABCB1 (rs2032582; rs1045642), MTHFR (rs1801131, rs1801133), HTR3A (rs1062613; rs1985242; rs1176713) and HTR3B (rs1176744).
-
Result: Enrolled were 121 JIA patients (82 girls: 39 boys) with a median age of 13.3 years (IQR: 11.3–15.1). The median MTX dose was 9.7 mg/m2/week (IQR: 9.0–10.9). The median MTX treatment duration prior to enrolment was 340 days (IQR: 142–766). The SNP analysis was available for 119 patients. MTX intolerance was associated with the genotype distribution of rs1801133 (MTHFR) (p = 0.02). There was no additive effect of the minor alleles for any of the selected SNPs, nor any significant haplotype associations.
Conclusion
-
Summary: MTX-induced nausea may be influenced by genetic polymorphisms in a MTX metabolizing enzyme (rs1801133; MTHFR).
-
Implications: Further analyses involving inclusion of larger cohorts are needed to understand the impact of SNPs on MTX-induced nausea in JIA.
Similar content being viewed by others
Findings
Background and hypothesis
Methotrexate (MTX) continues to have a key role in the treatment of children with juvenile idiopathic arthritis (JIA) [1,2,3]. MTX treatment is commonly associated with nausea. Large inter-individual variation exists in the level of MTX-induced nausea among children with JIA. From mild discomfort to a level of MTX-induced nausea that affects health-related quality of life to such an extent that premature termination is necessary [4,5,6]. Genetic factors are thought to influence the inter-individual variation in MTX-induced nausea.
MTX is mainly excreted through the kidneys, but the drug also undergoes enterohepatic circulation [7, 8]. The level of enterohepatic circulation is thought to be associated with the level of MTX-induced gastrointestinal side effects, including nausea [9]. Single nucleotide polymorphisms (SNPs) in genes encoding MTX transporter proteins may alter the function of these transporter proteins and hence affect the enterohepatic circulation of MTX and the level of MTX-induced gastrointestinal side effects [10,11,12,13] (Fig. 1).
MTX inhibits folate pathway enzymes including methylentetrahydrofolatereductase (MTHFR) [14]. This leads to an altered folate homeostasis and specifically the reduced MTHFR activity causes accumulation of homocysteine (Fig. 2), which has been hypothesized to be associated with MTX-induced gastrointestinal adverse events [15]. Specific SNPs in the gene encoding MTHFR have been shown to cause a decreased enzyme-activity [16, 17] and studies in smaller JIA cohorts have indicated that these SNPs may be associated with MTX-induced adverse effects [18, 19].
Illustration of the intracellular effects of methotrexate-polyglutamates including the role of the MTHFR enzyme. dUMP; deoxyuridine monophosphate. dTMP; deoxythymidine monophosphate. TYMS; thymidylate synthase. MTX-PG; methotrexate polyglutamates. DHFR; dihydrofolate reductase. DHF; dihydrofolate. THF; tetrahydrofolate. 5-CH3-THF; 5-methyltetrahydrofolate. 5,10-CH2-THF; 5,10-methylenetetrahydrofolate. MTHFR; methylenetetrahydrofolate reductase. ATIC; AICAR formyltransferase. AICAR; 5-aminoimidazole 4-carboxamide ribonucleotide. FAICAR; 10-formyl AICAR. IMP; inosine monophosphate. AMP; adenosine monophosphate
Nausea-inducing medicine can activate the 5-hydroxytryptamine type 3 (5-HT3)-receptor placed in the gastrointestinal tract and the medulla oblongata, which causes a signal to be transmitted to the emetic center in the central nervous system [20, 21] (Fig. 3). Studies have shown that SNPs in HTR3A and HTR3B encoding subunits of the 5-HT3-receptor are associated with impaired receptor function and nausea in adult cancer patients [22, 23]. Hence children with SNPs affecting the function of these genes may be more prone to experience MTX-induced nausea.
The objective for this study was to investigate whether MTX-induced nausea was associated with selected SNPs in candidate genes encoding MTX transporter proteins (SLCO1B1, SLCO1B3, SLC19A1, ABCB1, ABCC2), the MTHFR enzyme (MTHFR) and the 5-HT3-receptor (HTR3A, HTR3B), in children with JIA.
Methods
Study population
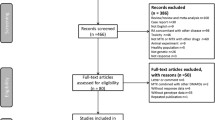
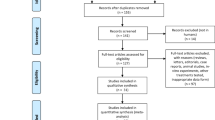
The study population was composed of children diagnosed with JIA according to the International League of Associations for Rheumatology criteria, treated with methotrexate and aged nine years or above. Children were excluded if cognitively impaired or not fluent in Danish (Fig. 4). All children were followed at our pediatric rheumatology outpatient clinics and enrolled from December 2013 until July 2016.
Phenotype
MTX-induced nausea was determined using two outcome measures previously described [24]: a Danish adaption of the methotrexate intolerance severity score (MISS) [6] and a nausea diary. The outcome measures were developed in electronic versions. The MISS was completed on the day of enrolment by the patients’ parents and they were instructed to focus on their child’s current state of MTX intolerance. The nausea diary was intended to be completed daily by the children for 28 consecutive days following enrolment. It consisted of categorically structured questions covering whether the child had experienced nausea and if so, its timely relation to MTX.
Phenotypes were established based on the two outcome measures. As defined by the MISS development paper [6], a child was categorised as MTXintolerant if the MISS total score was six or above with at least one point for an anticipatory and/or associative and/or behavioral symptom; otherwise categorised as MTXtolerant. The phenotype “MTX-nausea” was defined based on the nausea diary. It included children with a nausea diary illustrating a nausea pattern timely related to MTX administration, i.e. the child had to report a clear coherent pattern of nausea on the day of MTX administration and/or the day after; and at least have completed seven diary entries.
DNA extraction
On the day of enrolment a blood sample was drawn for the SNP analysis. DNA was extracted from EDTA blood by salt precipitation with triton lysis buffer (Tris 10 mM, MgCl2 5 mM, Triton X-100 1% and Sucrose 0.32 M), nucleus lysis buffer (NaCl 75 mM and EDTA 24 mM), saturated NaCl (6 M), isopropanol and Te-buffer (Tris-HCl 10 mM, EDTA 1 mM, pH 7.5). DNA concentration and purity was measured by a Nanodrop spectrophotometer.
SNP analysis
For the SNP analysis an amplicon with the specific SNP was created, PCR amplified and then sequenced by Sanger Sequencing. The analysis was performed at Eurofins Genomics using BigDye terminator cycle sequencing chemistry (version 3.1/3.0; Thermo Fisher Scientific), peqStar 96 HPL (PEQLAB Biotechnologie GMBH) and/or GeneTouch (Biozym Scientific GmbH) thermal cyclers, ABI3730xl capillary sequencers (Thermo Fisher Scientific) and GoTaq HotStart Green MasterMix (Promega). The quality scoring of the sequences was based on the quality value at the base-position, the peak-appearance, the overall signal intensity and quality of the sequence, the relation of forward and reverse sequence, and the identification of unspecific/artificial background signals or secondary PCR products and their subsequent signal. The quality scoring was done manually by Eurofins Genomics.
SNP selection
Within the specified candidate genes SNPs were selected based on clinical and genetic criteria. The SNPs should preferably have been proven clinically significant for gastrointestinal intolerance or toxicity, or proven phenotypically significant in animal studies. Further, they should have been studied in a relevant study population, have a reasonable minor allele frequency, a functional significance and preferably cause an amino acid change. The GTExportal.org was used to see whether the relevant gene section is expressed in relevant tissue. SNPsnap Broad Institute was used to see whether SNPs were in linkage disequilibrium.
Statistics
All statistical analyses were performed in STATA-13, PLINK 1.9 or haplo.stats R-package. Data on demographics and MTX treatment were analysed with Wilcoxon rank-sum test, chi-squared test/Fisher’s exact test or Student’s T-test, where applicable. The associations between SNP genotypes and phenotypes were analysed with chi-squared test/Fisher’s Exact test in STATA-13. Logistic regression (PLINK 1.9) was used to analyze the additive effects of the minor alleles. The analyses were filtered for Hardy-Weinberg equilibrium and took into account whether SNPs had more than two possible alleles present in the study population. Haplotype based association tests were performed for the two phenotypes – the method implemented in the R-package haplo.stats (https://CRAN.R-project.org/package=haplo.stats) and described by Schaid et al. [25] was used. The method allowed missing alleles and was developed for unrelated subjects, marker-alleles with unknown phase and ambiguous haplotypes. A progressive insertion algorithm was used to compute maximum likelihood estimates of haplotype probabilities. Additive models were chosen for all datasets.
Results
The study population encompassed 121 patients. Figure 4 illustrates a flowchart of the exclusion and enrolment of patients. There was missing data for the two outcome measures due to patients or parents never completing an entry, despite having consented to participation. Due to laboratory problems at enrolment SNP data was missing for two patients.
Demographics, MTX details
Table 1 shows demographic data, details of the MTX treatment and the distribution of children within the phenotype subgroups. The nausea diary was completed at least one day by 100/121 (83%) of the children. The 77 children who had completed at least seven diary entries were included in the genetic analysis for this phenotype. A total of 8/77 (10%) of the children reported no nausea in all their diary entries.
All patients were treated with folic acid supplement (5 mg) the day after MTX administration. Antiemetic medicine was prescribed for 41/121 (34%) of the children at enrolment.
SNPs in MTX transporter proteins
The selected SNPs within the candidate genes were: SLCO1B1 (rs4149056; rs4149081), SLCO1B3 (rs2117032), SLC19A1 (rs1051266), ABCC2 (rs2273697; rs3740066; rs717620), ABCB1 (rs2032582; rs1045642). No significant associations were found between the genotype distribution for these SNPs and the phenotype subgroups (Table 2), nor any significant additive effect of the minor alleles on either of the two phenotypes (MTXintolerant or “MTX-nausea”) (Table 3 and Table 4). No significant haplotype associations with either phenotype were found (Table 5 and Table 6).
SNPs in the MTHFR enzyme
The selected SNPs were rs1801131 and rs1801133. There was a significant association between the genotype distribution for rs1801133 and the parent-assessed phenotype (the MISS) (p = 0.02); but not for the child-reported phenotype (the nausea diary) (Table 2). There was no significant additive effect of the minor alleles on either phenotype for the two SNPs (Table 3 and Table 4), or any significant haplotype associations (Table 5 and Table 6).
SNPs in the 5-HT3-receptor
The selected SNPs within the candidate genes were: HTR3A (rs1062613; rs1985242; rs1176713) and HTR3B (rs1176744). No significant associations were found between the genotype distribution and the phenotype subgroups (Table 2). None of these SNPs were significantly associated with the two phenotypes (“MTX-nausea” or MTXintolerant) with regard to an additive effect of the minor alleles (Table 3 and Table 4). No significant haplotype associations with either phenotype were found (Table 5 and Table 6).
Conclusion
MTX-induced nausea may be influenced by SNPs in the MTHFR enzyme (rs1801133). Our data does not support an association between MTX-induced nausea and the remaining selected SNPs in genes encoding MTX-transporter proteins or the 5-HT3 nausea receptor.
Availability of data and materials
Authors can confirm that all relevant data are included in the article and its supplementary information files. The public availability of the data that support the findings of this study are restricted by the license of the Danish Data protection agency (1–16–02-429-15).
Abbreviations
- JIA:
-
Juvenile idiopathic arthritis
- SNPs:
-
Single nucleotide polymorphisms
- MISS:
-
Methotrexate intolerance severity score
- MTX:
-
Methotrexate
- MTHFR:
-
Methylentetrahydrofolatereductase
- 5-HT3 :
-
5-hydroxytryptamine type 3
References
Giannini EH, Brewer EJ, Kuzmina N, Shaikov A, Maximov A, Vorontsov I, et al. Methotrexate in resistant juvenile rheumatoid arthritis. Results of the U.S.a.-U.S.S.R. double-blind, placebo-controlled trial. The pediatric rheumatology collaborative study group and the cooperative Children's study group. N Engl J Med. 1992;326(16):1043–9. https://doi.org/10.1056/NEJM199204163261602.
Beukelman T, Patkar NM, Saag KG, Tolleson-Rinehart S, Cron RQ, DeWitt EM, et al. 2011 American College of Rheumatology recommendations for the treatment of juvenile idiopathic arthritis: initiation and safety monitoring of therapeutic agents for the treatment of arthritis and systemic features. Arthritis Care Res (Hoboken). 2011;63(4):465–82. https://doi.org/10.1002/acr.20460.
Ruperto N, Murray KJ, Gerloni V, Wulffraat N, de Oliveira SK, Falcini F, et al. A randomized trial of parenteral methotrexate comparing an intermediate dose with a higher dose in children with juvenile idiopathic arthritis who failed to respond to standard doses of methotrexate. Arthritis Rheum. 2004;50(7):2191–201. https://doi.org/10.1002/art.20288.
Brunner HI, Johnson AL, Barron AC, Passo MH, Griffin TA, Graham TB, et al. Gastrointestinal symptoms and their association with health-related quality of life of children with juvenile rheumatoid arthritis: validation of a gastrointestinal symptom questionnaire. J Clin Rheumatol. 2005;11(4):194–204. https://doi.org/10.1097/01.rhu.0000173616.81928.44.
Patil P, Parker RA, Rawcliffe C, Olaleye A, Moore S, Daly N, et al. Methotrexate-induced nausea and vomiting in adolescent and young adult patients. Clin Rheumatol. 2014;33(3):403–7. https://doi.org/10.1007/s10067-013-2389-x.
Bulatovic M, Heijstek MW, Verkaaik M, van Dijkhuizen EH, Armbrust W, Hoppenreijs EP, et al. High prevalence of methotrexate intolerance in juvenile idiopathic arthritis: development and validation of a methotrexate intolerance severity score. Arthritis Rheum. 2011;63(7):2007–13. https://doi.org/10.1002/art.30367.
Grim J, Chladek J, Martinkova J. Pharmacokinetics and pharmacodynamics of methotrexate in non-neoplastic diseases. Clin Pharmacokinet. 2003;42(2):139–51. https://doi.org/10.2165/00003088-200342020-00003.
Mikkelsen TS, Thorn CF, Yang JJ, Ulrich CM, French D, Zaza G, et al. PharmGKB summary: methotrexate pathway. Pharmacogenet Genomics. 2011;21(10):679–86. https://doi.org/10.1097/FPC.0b013e328343dd93.
Kato Y, Suzuki H, Sugiyama Y. Toxicological implications of hepatobiliary transporters. Toxicology. 2002;181–182:287–90.
Rau T, Erney B, Gores R, Eschenhagen T, Beck J, Langer T. High-dose methotrexate in pediatric acute lymphoblastic leukemia: impact of ABCC2 polymorphisms on plasma concentrations. Clin Pharmacol Ther. 2006;80(5):468–76. https://doi.org/10.1016/j.clpt.2006.08.012.
Trevino LR, Shimasaki N, Yang W, Panetta JC, Cheng C, Pei D, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27(35):5972–8. https://doi.org/10.1200/JCO.2008.20.4156.
Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem. 2001;276(38):35669–75. https://doi.org/10.1074/jbc.M103792200.
Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63(1):157–81. https://doi.org/10.1124/pr.110.002857.
Cronstein BN. The mechanism of action of methotrexate. Rheum Dis Clin N Am. 1997;23(4):739–55. https://doi.org/10.1016/S0889-857X(05)70358-6.
Ranganathan P, McLeod HL. Methotrexate pharmacogenetics: the first step toward individualized therapy in rheumatoid arthritis. Arthritis Rheum. 2006;54(5):1366–77. https://doi.org/10.1002/art.21762.
Schmiegelow K. Advances in individual prediction of methotrexate toxicity: a review. Br J Haematol. 2009;146(5):489–503. https://doi.org/10.1111/j.1365-2141.2009.07765.x.
Lopez-Lopez E, Martin-Guerrero I, Ballesteros J, Pinan MA, Garcia-Miguel P, Navajas A, et al. Polymorphisms of the SLCO1B1 gene predict methotrexate-related toxicity in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2011;57(4):612–9. https://doi.org/10.1002/pbc.23074.
Schmeling H, Biber D, Heins S, Horneff G. Influence of methylenetetrahydrofolate reductase polymorphisms on efficacy and toxicity of methotrexate in patients with juvenile idiopathic arthritis. J Rheumatol. 2005;32(9):1832–6.
Tukova J, Chladek J, Hroch M, Nemcova D, Hoza J, Dolezalova P. 677TT genotype is associated with elevated risk of methotrexate (MTX) toxicity in juvenile idiopathic arthritis: treatment outcome, erythrocyte concentrations of MTX and folates, and MTHFR polymorphisms. J Rheumatol. 2010;37(10):2180–6. https://doi.org/10.3899/jrheum.091427.
Chandrakantan A, Glass PS. Multimodal therapies for postoperative nausea and vomiting, and pain. Br J Anaesth. 2011;107(Suppl 1):i27–40. https://doi.org/10.1093/bja/aer358.
Wood GJ, Shega JW, Lynch B, Von Roenn JH. Management of intractable nausea and vomiting in patients at the end of life: "I was feeling nauseous all of the time . . . nothing was working". JAMA. 2007;298(10):1196–1207, Management of Intractable Nausea and Vomiting in Patients at the End of Life, DOI: https://doi.org/10.1001/jama.298.10.1196.
Laugsand EA, Fladvad T, Skorpen F, Maltoni M, Kaasa S, Fayers P, et al. Clinical and genetic factors associated with nausea and vomiting in cancer patients receiving opioids. Eur J Cancer. 2011;47(11):1682–91. https://doi.org/10.1016/j.ejca.2011.04.014.
Krzywkowski K, Jensen AA, Connolly CN, Brauner-Osborne H. Naturally occurring variations in the human 5-HT3A gene profoundly impact 5-HT3 receptor function and expression. Pharmacogenet Genomics. 2007;17(4):255–66. https://doi.org/10.1097/FPC.0b013e3280117269.
Kyvsgaard N, Thastum M, Mikkelsen TS, Christensen AE, Herlin T. Coping strategies and anxiety in association with methotrexate-induced nausea in juvenile idiopathic arthritis. Rheumatol Int. 2020;40(4):591–8. https://doi.org/10.1007/s00296-020-04520-9.
Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. https://doi.org/10.1086/338688.
Acknowledgements
Acknowledgements to Jane Hvarregaard Christensen for her contribution to the study through her genetic expertise; acknowledgements to Jane H. Knudsen and Tina Hindkjær for their contribution to the study through their laboratory work.
Funding
This study was supported by the Danish Childhood Cancer Foundation, the Danish Rheumatism Association and the Arvid Nilsson’ Foundation.
Author information
Authors and Affiliations
Contributions
NK, TSM, TJC, TH have made substantial contributions to the conception/design of the work. All authors have made substantial contributions to the acquisition of data, analysis and interpretation of data. NK compiled the first draft of the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was conducted in accordance with the declaration of Helsinki. All participating families provided written informed consent prior to enrolment and the Central Denmark Region’s research ethics committee approved the study (M-20110160). The study was observational and registered on clinicaltrials.gov (NCT02528435).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kyvsgaard, N., Mikkelsen, T.S., Als, T.D. et al. Single nucleotide polymorphisms associated with methotrexate-induced nausea in juvenile idiopathic arthritis. Pediatr Rheumatol 19, 51 (2021). https://doi.org/10.1186/s12969-021-00539-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12969-021-00539-9