Abstract
Background
Mitral annular plane systolic excursion (MAPSE) and left ventricular (LV) early diastolic velocity (e’) are key metrics of systolic and diastolic function, but not often measured by cardiovascular magnetic resonance (CMR). Its derivation is possible with manual, precise annotation of the mitral valve (MV) insertion points along the cardiac cycle in both two and four-chamber long-axis cines, but this process is highly time-consuming, laborious, and prone to errors. A fully automated, consistent, fast, and accurate method for MV plane tracking is lacking. In this study, we propose MVnet, a deep learning approach for MV point localization and tracking capable of deriving such clinical metrics comparable to human expert-level performance, and validated it in a multi-vendor, multi-center clinical population.
Methods
The proposed pipeline first performs a coarse MV point annotation in a given cine accurately enough to apply an automated linear transformation task, which standardizes the size, cropping, resolution, and heart orientation, and second, tracks the MV points with high accuracy. The model was trained and evaluated on 38,854 cine images from 703 patients with diverse cardiovascular conditions, scanned on equipment from 3 main vendors, 16 centers, and 7 countries, and manually annotated by 10 observers. Agreement was assessed by the intra-class correlation coefficient (ICC) for both clinical metrics and by the distance error in the MV plane displacement. For inter-observer variability analysis, an additional pair of observers performed manual annotations in a randomly chosen set of 50 patients.
Results
MVnet achieved a fast segmentation (<1 s/cine) with excellent ICCs of 0.94 (MAPSE) and 0.93 (LV e’) and a MV plane tracking error of −0.10 ± 0.97 mm. In a similar manner, the inter-observer variability analysis yielded ICCs of 0.95 and 0.89 and a tracking error of −0.15 ± 1.18 mm, respectively.
Conclusion
A dual-stage deep learning approach for automated annotation of MV points for systolic and diastolic evaluation in CMR long-axis cine images was developed. The method is able to carefully track these points with high accuracy and in a timely manner. This will improve the feasibility of CMR methods which rely on valve tracking and increase their utility in a clinical setting.
Similar content being viewed by others
Background
The mitral valve (MV) is a fibrous region that separates the left ventricle (LV) and the left atrium with two leaflets. In the normal heart, the MV remains closed during systole and the MV plane rapidly descends with contraction of the ventricles; in early diastole the MV opens, and the MV plane quickly springs back to an equilibrium plane where it pauses during diastasis, and then ascends further during the left atrial kick in late diastole. The analysis of MV plane motion provides structural and functional systolic and diastolic information [1]. Its measurement yields peak displacement of the plane during systole, known as mitral annular plane systolic excursion (MAPSE), and the LV early diastolic velocity, known as LV e’, which is itself a key metric of diastolic function in echocardiography [2].
Cardiovascular magnetic resonance (CMR) is a reproducible imaging modality and is considered the reference standard for cardiac volume assessment. Its accuracy and reliability hold promise for serial examinations of MAPSE and LV e’, as already reported in our previous work [3]. These metrics were obtained by tracking the MV insertion points in every frame in long-axis cine images. MV plane tracking has also been used to enable slice-following for assessment of valvular flow with a phase-contrast sequence, either retrospectively [4] or prospectively [5], where it allows an evaluation of mitral regurgitation, which would not be possible without valve tracking. The MV plane location can assist any automated segmentation of the LV or left atrium, as it demarcates these chambers [6], and its tracking can provide an estimate of global longitudinal strain [7]. Finally, MV plane dynamics could be useful in providing information on the timing of the cardiac rest-periods, which is important in CMR [8,9,10].
Even with significant improvements in semi-automated tracking methods of the MV points in cine CMR [11,12,13,14,15,16], and validation of clinical metrics against echocardiography [3, 16], MV plane tracking still requires a manual initialization and refinement. A fully automated, fast and accurate method for tracking the MV points using standard clinical CMR images is lacking. The rapid evolving field of deep learning has great potential to provide such method. In CMR deep learning applications [17], and deep learning in general, it is strongly suggested to have a large cohort of training data coming from different centers, vendors, pathologies, and manually labeled by different experts. Such diversity in the data leads to a consistent, robust method to perform a determined task.
In this study, we develop MVnet, a dual-stage deep learning approach for MV tracking using residual neural networks, trained and tested in a multi-center, multi-vendor population of 703 patients, with a wide range of pathologies, and manually labeled by 10 experts. Additionally, we show the importance of using a rich training dataset by structurally analyzing two main scenarios with direct impact on the learning and application: a single-center, single-vendor, single-expert dataset compared to a multi-center, multi-vendor, multi-expert dataset. We also evaluate the derived clinical parameters (MAPSE and LV e’) in contrast with their counterpart by experts. Finally, with this dual-stage pipeline, we present technical novelty by applying an automated linear transformation to the images after the first stage, which substantially reduces variance and boosts performance in accuracy.
Methods
Imaging data
A multi-center, multi-vendor population of 703 subjects (226 females, 51±19 years old) for diverse clinical indications were retrospectively enrolled, as part of IRB approved chart-review studies. The reported pathologies (Table 1) included subjects with myocardial infarction (n = 169), chronic heart failure (n = 130), arrhythmia - mostly atrial fibrillation (n = 63), heart failure with reduced LV ejection fraction (n = 54), endurance athletes (n = 39), pulmonary arterial hypertension (n = 25), atrial septal defect (n = 19), hypertrophic cardiomyopathy (n = 14), other cardiac diseases (n = 13), sarcoid (n = 6), myocarditis (n = 6), and healthy volunteers of all ages (n = 165). All subjects were scanned on a 1.5T (n = 661) or a 3T (n = 42) conventional clinical CMR scanners from Philips Healthcare (Best, the Netherlands; n = 419), Siemens Healthineers (Erlangen, Germany; n = 250), and General Electric Healthcare (Chicago, Illinois, USA; n = 34). Inclusions were performed in 16 different centers and 7 countries, and included standard two-chamber and four-chamber long-axis cine exams, according to clinical practice. Typical image parameters were breath-hold with repetition time of 2.8 to 3.0 ms, echo time of 1.4 to 1.5 ms, and flip angle of 50 to 60°. Imaging data of both chamber views had a spatial resolution ranging from 1.3 \(\times\) 1.3 mm2 to 1.7 \(\times\) 1.7 mm2 (after zero-filling), slice thickness of 8 mm and typically 30 (25 to 50) temporal frames per cardiac cycle were reconstructed. The final dataset comprised a total of 38,854 images (703 sets of time-resolved images) analyzed in previous studies at Yale University (Yale dataset) [3, 6, 18, 19] and at Lund University (Lund dataset) [20,21,22,23,24,25,26,27,28,29,30,31,32,33].
Manual annotation
Using an analysis tool, freely available in the software Segment [34], 10 trained human observers with different backgrounds and levels of CMR experience, ranging from 4 to 20 years, performed these annotations in all temporal frames of 458 subjects and only at end-diastolic and end-systolic frames of 245 subjects (belonging to the Lund dataset). The experts placed the MV insertion points in two-chamber view data, as anterior and inferior points, and in four-chamber view data, as lateral and septal points, at each phase in the cardiac cycle, resulting in two points for each view in each image (Fig. 1a).
a MV point annotation illustration at end-diastole and end-systole in 2-chamber view, with anterior and inferior points, and 4-chamber view, with lateral and septal points, and b the clinical-metric derivation as an output of the time-resolved annotation. The MV displacement was measured as the average of the perpendicular distances from the MV initial plane, defined at end-diastole in each view, to every MV point at every temporal frame, and the MV velocity was measured as its time-derivative. MAPSE was calculated as the maximum displacement, and LV s’, e’ and a’ as the first, second and third global velocity peak. MV mitral valve, MAPSE mitral annular plane systolic excursion, LV left ventricle
With no society recommendations on how to annotate and track the MV points, the Yale and Lund datasets were annotated according to two different principles. A single observer at Yale University (Yale observer) placed the MV points in the intersection between the MV and the LV myocardium, whereas the 9 observers at Lund University (Lund observers) placed the MV points at the most basal part of the compact LV myocardium. Both ways yielded similar MV motions but with a different reference point. However, due to the inherent observer bias and the high number of Lund observers, these annotations presented more variability. Such differences in manual annotation are very common in deep learning applications.
Dual-stage residual neural network
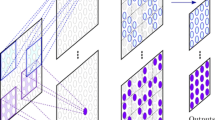
Based on our recent work to track the tricuspid valve [19] as proof of principle, we adapted and expanded our dual-stage residual neural network to track the MV. The proposed framework (Fig. 2) involved two stages for each chamber view. The first stage uses a trained network to track the MV points with sufficient accuracy to define the MV plane, and the second stage uses these points to perform a linear transformation on the original images to standardize the images regarding location and orientation of the valve plane. The second network then predicts the MV points in the automatically preprocessed image with higher accuracy. Each stage uses an artificial neural network with a residual framework of 50 layers, ResNet-50 [35], adapted to predict a series of four numbers (representing two pairs of coordinates \(\{x,y\}\)) on an individual grayscale cine image (Fig. 3).
MVnet pipeline. a The input cine images with an inherent clinical variability (size \(m \times n\), resolution, orientation and cropping) were fed to the proposed dual-stage residual neural network. b The first trained ResNet-50 produced coarse annotations, marked in circumferences representing acceptable accuracy, in every cine image in a fixed image size of 160 \(\times\) 160, which in turn, served to apply a c linear transformation to a standard spatial resolution of 0.75 mm, orientation and cropping around the mitral valve center for a size of 118 \(\times\) 162. d The second trained ResNet-50 used the transformed images to predict precise annotations, marked in circles representing higher accuracy, which were adjusted again to the original input image. These last two tasks could be done iteratively as indicated. e The output time-resolved coordinates were used to derive the mitral valve displacement and velocity curves
Adapted convolutional neural network architecture with a residual learning framework, ResNet50 [35], used in this study for automated mitral valve point tracking. With a given input image with size \(m \times n\), the network outputs two mitral valve points \(\{x_1, y_1, x_2, y_2\}\) in Cartesian coordinates
Accuracy of each model (MVnet) trained and evaluated on its own dataset, by the mean a Euclidean and b angular distance error, and the agreement with ICC in c MAPSE, d LV e’ and e MV displacement, stratified by the output of the first stage (stage 1), second stage (stage 1+2), and an iteration of the second stage (stage 1+2+2). For (a, b), each bar represents the mean, and error bar the standard deviation of each accuracy metric. For (c, d, e), each bar represents the ICC, and error bar the confidence interval (95%) of each accuracy metric. The output of the iteration (stage 1+2+2) achieved the best accuracy and was chosen for the proposed workflow. ICC intra-class correlation coefficient, MAPSE mitral annular plane systolic excursion, LV left ventricle, MV mitral valve
Accuracy of each model (MVnet) trained and evaluated on its own dataset, by the mean Euclidean distance error (first two columns) and angular distance error (third column), stratified by the output of the first stage (stage 1), second stage (stage 1+2), and an iteration of the second stage (stage 1+2+2). Accuracy assessed for 2Ch in its a anterior and b inferior point distance error, and c angular distance error; and for 4Ch in its d left ventricular lateral and e septal point distance error, and f angular distance error. Each bar represents the mean, and error bar the standard deviation of each accuracy metric. 2Ch two-chamber view, 4Ch four-chamber view
Accuracy heatmap of each model (MVnet) trained on each training set and evaluated on each test set by the mean a Euclidean and b angular distance error, and the agreement with ICC in c MV displacement, d MAPSE, and e LV e’. The output of \({{\text{MVnet}}_{\text{Mixed}}}\) consistently achieved the best accuracy and was chosen for the proposed model. ICC intra-class correlation coefficient, MV mitral valve, MAPSE mitral annular plane systolic excursion, LV left ventricle
Clinical-metric agreement of a MV displacement, b MAPSE and c LV e’ between an expert manual annotation and the automated method (\({{\text{MVnet}}_{\text{Mixed}}}\)). The first row of each analysis shows the regression plots whereas the second shows the Bland-Altman plots. In each scatter plot the black line denotes the identity line, whereas in each Bland-Altman plot, the red line denotes the mean difference (bias) and the two light dotted lines denote ± 1.96 standard deviations from the mean. MV mitral valve, MAPSE mitral annular plane systolic excursion, LV left ventricle
Clinical-metric agreement of a MV displacement, b MAPSE and c LV e’ between an expert manual annotation (observer 1) and a pair of second observers (observer 2). One observer from the Yale dataset and another from the Lund dataset annotated 25 subjects from each test set. The first row of each analysis shows the regression plots whereas the second shows the Bland-Altman plots. In each scatter plot the black line denotes the identity line, whereas in each Bland-Altman plot, the red line denotes the mean difference (bias) and the two light dotted lines denote ± 1.96 standard deviations from the mean. MV mitral valve, MAPSE mitral annular plane systolic excursion, LV left ventricle
Stage 1
The first stage only involves the use of one network to perform a coarse annotation in all temporal frames. The network was trained on manually annotated images resized to 160 \(\times\) 160 with cubic interpolation, leading to anisotropic pixel dimensions. This initial coarse annotation serves to localize the MV plane and follow its motion.
Stage 2
The second stage uses the output coordinates of the previous stage to apply a linear transformation task, i.e., standardize the cine, by (i) interpolating each image to a spatial isotropic resolution of 0.75 mm, (ii) rotating each image such that the MV is oriented horizontally with the apex pointing down and with the anterior and lateral points placed on the left and the inferior and septal points placed on the right, for two-chamber and four-chamber views, respectively, and (iii) cropping each image around the MV center for a size of 118 \(\times\) 162.
Pipeline
As an overview, a given input cine of a chamber view with different parameters (Fig. 2a) is fed into the first stage with a fixed size for the network to read (Fig. 2b). The first trained network (Fig. 3) outputs the predicted points with acceptable accuracy. The coarse annotation on the first temporal frame is used as a reference to determine the orientation and centering tasks, i.e., to horizontally orient the MV in the center, whereas the coarse annotation on the remaining temporal frames determine the motion direction, i.e., to ensure the apex points down. The automated linear transformation task from the second stage standardizes the spatial resolution, the heart orientation and positioning and the type of cropping (Fig. 2c). The second network (Fig. 3), trained on linearly transformed images, is then used to track the MV points with high accuracy (Fig. 2d). These new predicted points are readjusted, with an inverse standardization transformation, to match the original input cine images (Fig. 2e). This stage can be performed in an iterative manner as indicated, i.e, the points predicted by the second stage can initialize again the second stage (linear transformation task and second network) to yield a more accurate annotation.
Network training
Both networks were trained with the same process but independently for each stage and chamber view. Transfer learning for weights initialization was applied to reduce convergence time and aid the learning process, using a ResNet-50 pretrained on more than one million images from the ImageNet database [36] for a classification task into 1000 different categories of objects and animals photographs. Standard data preparation involved pixel distribution normalization by the median and interquartile range to ensure generalizability [37]. Training data was augmented 10 times by scaling ±10%, rotating ±10\(^{\circ }\) and translating ±3 pixels to add more inherent variability in the first stage, and to compensate for any error introduced from the first stage to the second stage, i.e., a slight misalignment of the MV plane center from the ground truth. The mean square error loss function was optimized by the Adam method [38] with a learning rate of \(1\times 10^{-4}\), for 20 epochs and mini-batch size of 8. The pipelines were developed on MATLAB R2019b (Mathworks, Natick, MA) with a NVIDIA Titan RTX GPU.
Clinical metric derivation
The MV plane displacement was calculated for each chamber view as the average perpendicular distance of the MV points to the initial plane set in end-diastole (Fig. 1b). The resultant MV plane displacement was measured as the average from both chamber views. MAPSE was derived from the maximal MV displacement and LV e’ was the second global peak of the time-derivative of the displacement curve.
Evaluation
Data variability analysis
Both Yale and Lund datasets were trained and tested separately and collectively to assess the influence of employing a single-center, single-vendor, single-expert dataset (Yale) and a multi-center, multi-vendor, multi-expert dataset (Lund). This analysis was performed in three different pipelines: \({{\text{MVnet}}_{\text{Yale}}}\), \({{\text{MVnet}}_{\text{Lund}}}\) and \({{\text{MVnet}}_{\text{Mixed}}}\), for the Yale dataset, Lund dataset, and both mixed, respectively. Each MVnet comprised of 4 networks in total, i.e., 2 networks for the first and second stage of the two-chamber view and other 2 networks for both stages of the four-chamber view. For \({{\text{MVnet}}_{\text{Yale}}}\), the training and testing sets were partitioned into 6948 images (118 subjects) and 1886 images (32 subjects), respectively. For \({{\text{MVnet}}_{\text{Lund}}}\), the training and testing sets were partitioned into 26,072 images (118 subjects) and 3948 images (111 subjects), respectively. For \({{\text{MVnet}}_{\text{Mixed}}}\), the same distribution set for each dataset was used and mixed. Such distributions (Table 2) were randomly performed with a constraint to have an homogeneous representation of the reported cardiovascular diseases.
Spatial annotation accuracy
The test set of each MVnet was evaluated against manual annotation. The spatial annotation error was measured in each chamber view with: (i) the Euclidean distance, which measured in millimeters the distance error between the ground-truth and predicted annotations, (ii) the angular distance, which measured in degrees the inner intersection angle of the ground-truth and automated planes defined by both MV points, and (iii) the MV displacement, which measured in millimeters the difference in the MV plane displacement in every temporal frame between the ground-truth and automated displacement curves. All metrics were calculated comparing the ground-truth with the predicted annotations on the original input images.
Clinical-metric accuracy
Clinical metric (MAPSE and LV e’) comparisons were performed using linear regression analysis, Bland-Altman plots, and the intra-class correlation coefficient (ICC) between the automated and manual measures. As 47 subjects (out of 111) from the Lund test set were only annotated at end-diastole and end-systole, minimum amount of temporal frames required for MAPSE, LV e’ comparisons were not performed in that subset. The threshold for statistical significance was considered to be p<0.05 for this study.
Dual-stage influence
Both spatial annotation and clinical-metric accuracy evaluations were performed for the results of the first stage (stage 1), both stages (stage 1 + 2) and an additional iteration (stage 1 + 2 + 2), to show the influence of the additional stage (Fig. 2c, d).
Inter-observer variability analysis
For inter-observer variability analysis, an additional pair of observers performed manual annotations in a randomly chosen of 50 subjects. Specifically, a second observer of Yale dataset (Yale observer 2) and Lund dataset (Lund observer 2) manually annotated a subset of 25 subjects each on its corresponding dataset and the same evaluation was assessed.
Results
Implementation
MVnet was implemented in the medical image analysis software Segment v3.1 R8109 [34] (http://segment.heiberg.se), which is freely available for research purposes, and uploaded to https://github.com/ra-gonzales/MVnet. Total training time, including each stage and chamber view, took 108, 260 and 420 hours for \({{\text{MVnet}}_{\text{Yale}}}\), \({{\text{MVnet}}_{\text{Lund}}}\) and \({{\text{MVnet}}_{\text{Mixed}}}\), respectively. For each \({\text{MVnet}}\), on the GPU, testing time took 5.2 seconds per patient, whereas on a CPU, it took 10.8 seconds per patient, including data I/O time, compared to an average manual annotation time from 8 to 20 minutes. Batch processing reduces the average time to under 1 second on the GPU.
Dual-stage influence
The annotation accuracy of each \(\text {MVnet}\) after the first stage (stage 1), the second stage (stage 1+2) and an iteration of the second stage (stage 1+2+2) is reported in Fig. 4, in terms of spatial annotation and clinical-metric accuracy, between every ground-truth and predicted measures of an \(\text {MVnet}\) with its corresponding test set.
In terms of Euclidean and angular distance agreement (Fig. 4a, b), the mean percentage error of both metrics from the first to the second stage decreased 34%, 14%, and 10% for \({{\text{MVnet}}_{\text {Yale}}}\), \({{\text{MVnet}}_{\text{Lund}}}\) and \({{\text{MVnet}}_{\text {Mixed}}}\), respectively. In a similar manner, the overall error from the first stage to the second iteration of the second stage decreased 41%, 15%, and 11%, respectively. Accuracy and agreement were consistently improved after each stage. No substantial differences were found in a specific MV point and chamber view (Fig. 5), meaning they all achieved a similar accuracy. However, the angular distance error on the four-chamber view was larger with a higher discordance in the septal point placement from the difference in the two annotation principles.
Regarding MV displacement, MAPSE and LV e’ agreement (Fig. 4c–e), with the addition of the second stage, the mean percentage error decreased 42%, 29%, and 11% for \({{\text{MVnet}}_{\text{Yale}}}\), \({{\text{MVnet}}_{\text{Lund}}}\) and \({{\text{MVnet}}_{\text{Mixed}}}\), respectively, whereas the iteration of the second stage reduced the initial error 39%, 32%, and 18%. This iteration improved the agreement with clinical metrics, except for \({{\text{MVnet}}_{\text{Yale}}}\) which showed a small reduction in agreement. Although the improvement of the iteration was moderate in the spatial annotation accuracy for \({{\text{MVnet}}_{\text{Lund}}}\) and \({{\text{MVnet}}_{\text{Mixed}}}\), the clinical-metric accuracy was further improved.
Dataset variability influence
With the output of each \(\text {MVnet}\) considered to be the predictions after an iteration of the second stage (stage 1+2+2), the accuracy of every model evaluated on every test set (the Yale, Lund and mixed test sets) is reported as a heatmap for each metric in Fig. 6, where the best performance among the models is highlighted in blue and the worst in red. Although both \({{\text{MVnet}}_{\text{Yale}}}\) and \({{\text{MVnet}}_{\text{Lund}}}\) performed well on their corresponding datasets, with generally superior performance for Lund, the accuracy was noticeably reduced when the Yale (or Lund) network was applied to Lund (or Yale) test dataset, with Lund (or Yale) annotations. Then there was a 2 to 3 times-fold error increase in Euclidean and angular distances, mainly as a result of difference in annotation pattern. The MV displacement metric, however, achieved a better agreement in this scenario, but still with lower performance.
While \({{\text{MVnet}}_{\text {Yale}}}\) achieved the lowest Euclidean distance error (although trained on less data), its error on the Lund test set was 3.3 times higher. Vice versa, \({{\text{MVnet}}_{\text{Lund}}}\) on the Yale test set was also notably higher (2.4 times). Similarly, the clinical-metric agreement of a model tested in a different test set was markedly decreased. In the case of LV e’, \({{\text{MVnet}}_{\text{Lund}}}\) failed on predicting the same clinical values as the manual measures with an ICC of 0.47. In contrast, \({{\text{MVnet}}_{\text{Mixed}}}\) performed with the same consistency, and overall it achieved a better, more robust agreement with both groups of human experts.
Clinical-metric accuracy
Choosing \({{\text{MVnet}}_{\text{Mixed}}}\) as the proposed pipeline, the accuracy of clinical metrics for MAPSE and LV e’ of the automated method compared against the manual metrics, evaluated in the mixed test set, are shown in Table 3. The model estimated both metrics with excellent agreement with a mean error of −0.2±1.3 mm (ICC = 0.94) and 0.0±1.5 cm/s (ICC = 0.93), for MAPSE and LV e’, respectively. The regression and Bland-Altman plots for the MV parameters between the automated and manual measurements are presented in Fig. 7, where an excellent correlation and good agreement were observed for each of the three parameters, including the MV displacement. All reported correlation values are significant (p<0.0001).
Inter-observer variability analysis
The inter-observer clinical-metric agreement is shown in Table 4. The results were on par with the automated predictions, with a mean error of −0.3±1.2 mm (ICC = 0.95) and 0.3±1.7 cm/s (ICC = 0.89), for MAPSE and LV e’, respectively. In a similar manner, the regression and Bland-Altman plots for the clinical parameters between the second and first group of observers are presented in Fig. 8. All reported correlation values are also significant (p<0.0001).
Spatial annotation accuracy comparison
The spatial annotation agreement of the automated annotations by \({{\text{MVnet}}_{\text{Mixed}}}\) against manual annotations as well as the inter-observer spatial variability are presented in Table 5. Interestingly, while the Euclidean and angular distance errors seem to lower the pipeline performance, the automated reproducibility of clinical metrics is very high resulting in a very low MV displacement error. This shows that tracking the motion is more reproducible, and more relevant, than tracking the spatial location of each individual point. Therefore, the individual distance errors are not necessarily a good metric for evaluating the performance of a valve plane movement and may be misleading when the accuracy of the annotation model is very high. This discrepancy was also present in the studied inter-observer variability analysis as the Yale observer 2 yielded a Euclidean distance error of 2.7 ± 2.6 mm against ground truth, whereas the error of the Lund observer 2 was 3.9 ± 3.0 mm. Although this difference may flag a potential pitfall in the manual annotation, the clinical-metric agreement showed the opposite as the latter achieved an average ICC = 0.97, whereas the former an average ICC = 0.82, indicating that annotation consistency along temporal frames prevails above a specific annotation pattern.
Additional movie files demonstrated MV tracking in cines from the Yale test set (Additional file 1) and in the Lund data set (Additional file 2), using the automated annotations by \({{\text{MVnet}}_{\text{Mixed}}}\). Additional files also show further analysis of the inter-pipeline variability (Additional file 3) and clinical-metric agreement of LV s’ and LV a’, compared with the inter-observer variability (Additional file 4).
Discussion
In this work, we proposed a dual-stage residual learning framework, MVnet, for time-resolved annotation of the MV in two-chamber and four-chamber views from standard long-axis cine CMR images. The proposed method was fast, fully automated and showed excellent agreement with manual annotation by expert readers in term of valve points positioning as well as with the subsequently extracted LV function parameters. Tedious manual labor is not needed, reducing the processing time from 8 to 20 minutes to 5 or 1 second with batch processing. This enables fully-automated, accurate, fast, and reproducible assessment of LV function in clinical routine. Moreover, this method can be applied retrospectively to any two and four-chamber image acquisition, which are routinely acquired in a standard CMR exam. Additionally, we have systematically shown the advantages and pitfalls of using a single-center, single-vendor, single-expert dataset compared with a multi-center, multi-vendor, multi-expert dataset.
The technical contribution of this work includes the accuracy improvement provided by the second stage. While both networks were trained under the same domain and task, meaning each of them predicts two pairs of coordinates in a given image, the second network processes only highly standardized images, obtained by applying the proposed linear transformation. We showed how much each additional stage, up to one iteration (stage 1+2+2), improved the overall performance. While this assessment could be performed with more iterations (e.g., 1+2+2+2), the accuracy does not further improve, as evaluated in our proof-of-principle work [19]. This adoption of a dual-stage deep learning pipeline in biomedical applications has recently gained some interest to compensate for the technical limitation of one single model, even when trained with a large amount of data. For instance, some dual-stage pipelines with a segmentation task [39, 40] first localize the region of interest with a bounding box and then segment the bounded image, which improved the accuracy from single models. One limitation for an approach consisting of two different tasks is that an error for the localization task can hamper the performance. In our case, we employed the same annotation task for both stages, with a good accuracy on the first stage and an increased performance with the second one. The first stage is enriched by the large-scale study, from different centers, vendors and observers, and data augmentation to further increase the sample diversity [41] allowing model generalization [42]. This approach is commonly employed by one-stage deep learning applications where it is believed that data diversity will solve the image processing task. The specific technical contribution of our work is that we used the benefit of data diversity but also further increased the accuracy by using these results to standardize the images in a novel way.
Our proposed method achieved human-level performance with high robustness and consistency across centers, vendors and observers in a diverse range of conditions. The value of including images from different centers, vendors and conditions aids the generalization of the trained model through seeing all potential variations of the images in a real case scenario [42], whereas including different observers reduces the bias of a single observer [43]. Although this is not the first data diversity study in CMR, as another multi-vendor, multi-center study [44] also evaluated the incremental training strategy for a LV segmentation task with a thorough assessment, our work additionally assessed the annotation pattern diversity. We showed how a model trained on one center could yield a high accuracy in its own dataset but underperformed in datasets from different centers, to achieve generalizable multi-center development [37], and how a different annotation pattern generated discordance against another pattern, even with diverse training inputs.
We evaluated the performance against ground truth with a wide range of parameters including the Euclidean distance, angular distance, MV displacement, MAPSE and LV e’. We showed that the proposed pipeline yielded high reproducibility in a very demanding task with excellent ICC for MV displacement, MAPSE and LV e’, and how the Euclidean distance error may be misleading. This error discrepancy between training labels and clinical metrics has been noted by others. A recent learning-based approach for myocardial segmentation on T1 maps [45] achieved a near-perfect accuracy on estimating T1 values of LV myocardium, even while the segmentation accuracy only yielded a Dice similarity coefficient [46] of 0.85. Although this metric could be misleading, the clinical-metric agreement prevails over image processing performance, as the most important metric. In our study, we showed how a Euclidean distance error could be higher, but the MV plane displacement error was lower.
Limitations
One limitation of our study is the lack of consensus on how to annotate the MV points as a specific pattern would have homogenized the bias and yielded a lower error. However, our thorough analysis demonstrated that the difference in the annotation patterns did not hamper the performance but instead the MVnet reduced such biases and learned a consensual pattern from the diverse observers, confirming the value of multiple observers for a deep learning application [43]. Another limitation of our study is the missing benefit from incorporating a recurrent neural network architecture to learn spatiotemporal dependencies across the cardiac cycle instead learning from one temporal frame at a time. However, the technical contribution of this work relies on the automated image standardization algorithm to boost both image processing performance and clinical metric agreement, implementing this pipeline to a recurrent neural network architecture may also benefit its learning.
The clinical value of this work is that it provides an automated method for MV plane motion, including established metrics such as MAPSE and LV e’, and also utility for slice-following applications, automated cardiac rest-period identification, among others. Additionally, it does not need any added work in the clinical routine and the post-processing cost is negligible.
Conclusion
MVnet is a deep learning approach for automated delineation of MV points for MV plane displacement evaluation in CMR long-axis cine images. The method is able to track the MV points, accurately, rapidly and consistently. This will improve the feasibility of CMR methods which rely on valve tracking, such as measurement of e’, or slice-following phase-contrast, and increase their utility in a clinical setting.
Availability of data and materials
The implementation of MVnet will be made freely available in the software Segment (http://www.medviso.com/segment). Imaging data can not be shared due to data privacy consideration details. Other data in the paper will be made available upon reasonable request.
Abbreviations
- 2Ch:
-
2-Chamber
- 4Ch:
-
4-Chamber
- CMR:
-
Cardiovascular magnetic resonance
- e':
-
Early diastolic velocity
- ICC:
-
Intra-class correlation coefficient
- LV:
-
Left ventricle/left ventricular
- MAPSE:
-
Mitral annular plane systolic excursion
- MV:
-
Mitral valve
References
Carlsson M, Ugander M, Mosén H, Buhre T, Arheden H. Atrioventricular plane displacement is the major contributor to left ventricular pumping in healthy adults, athletes, and patients with dilated cardiomyopathy. Am J Physiol Heart Circ Physiol. 2007;292(3):1452–9. https://doi.org/10.1152/ajpheart.01148.2006.
Nagueh SF, Smiseth OA, Appleton CP, Byrd I. Benjamin F, Dokainish H, Edvardsen T, Flachskampf FA, Gillebert TC, Klein AL, Lancellotti P, Marino P, Oh JK, Popescu B, Waggoner AD. Recommendations for the evaluation of left ventricular diastolic function by echocardiography: an update from the american society of echocardiography and the European association of cardiovascular imaging. Eur Heart J 2016;17(12):1321–1360. https://doi.org/10.1093/ehjci/jew082
Seemann F, Baldassarre LA, Llanos-Chea F, Gonzales RA, Grunseich K, Hu C, Sugeng L, Meadows J, Heiberg E, Peters DC. Assessment of diastolic function and atrial remodeling by MRI - validation and correlation with echocardiography and filling pressure. Physiol Rep. 2018;6(17):13828. https://doi.org/10.14814/phy2.13828.
Roes SD, Hammer S, van der Geest RJ, Marsan NA, Bax JJ, Lamb HJ, Reiber JHC, de Roos A, Westenberg JJM. Flow assessment through four heart valves simultaneously using 3-dimensional 3-directional velocity-encoded magnetic resonance imaging with retrospective valve tracking in healthy volunteers and patients with valvular regurgitation. Invest Radiol. 2009;44:10. https://doi.org/10.1097/rli.0b013e3181ae99b5.
Seemann F, Heiberg E, Carlsson M, Gonzales RA, Baldassarre LA, Qiu M, Peters DC. Valvular imaging in the era of feature-tracking: a slice-following cardiac MR sequence to measure mitral flow. J Magn Reson Imag. 2020;51(5):1412–21. https://doi.org/10.1002/jmri.26971.
Gonzales RA, Seemann F, Lamy J, Arvidsson PM, Heiberg E, Murray V, Peters DC. Automated left atrial time-resolved segmentation in MRI long-axis cine images using active contours. BMC Med Imag. 2021;21(1):101. https://doi.org/10.1186/s12880-021-00630-3.
Leng S, Ge H, He J, Kong L, Yang Y, Yan F, Xiu J, Shan P, Zhao S, Tan R-S, Zhao X, Koh AS, Allen JC, Hausenloy DJ, Mintz GS, Zhong L, Pu J. Long-term prognostic value of cardiac MRI left atrial strain in ST-segment elevation myocardial infarction. Radiology. 2020;296(2):299–309. https://doi.org/10.1148/radiol.2020200176.
Plein S, Jones TR, Ridgway JP, Sivananthan MU. Three-dimensional coronary MR angiography performed with subject-specific cardiac acquisition windows and motion-adapted respiratory gating. Am J Roentgenol. 2003;180(2):505–12. https://doi.org/10.2214/ajr.180.2.1800505.
Jahnke C, Paetsch I, Nehrke K, Schnackenburg B, Bornstedt A, Gebker R, Fleck E, Nagel E. A new approach for rapid assessment of the cardiac rest period for coronary MRA. J Cardiovasc Magn Reson. 2005;7(2):395–9. https://doi.org/10.1081/jcmr-200053616.
Markus R, Hussain M, Batsis M, Zahr RA, Tandon A, Dyer A, Greil G. Velocity encoded cine imaging of mitral valve inflow: a novel method to determine cardiac rest periods in coronary magnetic resonance imaging. Pediatrics. 2019. https://doi.org/10.1542/peds.144.2_MeetingAbstract.335.
Maffessanti F, Gripari P, Pontone G, Andreini D, Bertella E, Mushtaq S, Tamborini G, Fusini L, Pepi M, Caiani EG. Three-dimensional dynamic assessment of tricuspid and mitral annuli using cardiovascular magnetic resonance. Eur Heart J. 2013;14(10):986–95. https://doi.org/10.1093/ehjci/jet004.
Wu V, Chyou JY, Chung S, Bhagavatula S, Axel L. Evaluation of diastolic function by three-dimensional volume tracking of the mitral annulus with cardiovascular magnetic resonance: comparison with tissue Doppler imaging. J Cardiovasc Magn Reson. 2014;16(1):71. https://doi.org/10.1186/s12968-014-0071-3.
Saba SG, Chung S, Bhagavatula S, Donnino R, Srichai MB, Saric M, Katz SD, Axel L. A novel and practical cardiovascular magnetic resonance method to quantify mitral annular excursion and recoil applied to hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2014;16(1):35. https://doi.org/10.1186/1532-429X-16-35.
Leng S, Zhao X-D, Huang F-Q, Wong J-I, Su B-Y, Allen JC, Kassab GS, Tan R-S, Zhong L. Automated quantitative assessment of cardiovascular magnetic resonance-derived atrioventricular junction velocities. Am J Physiol Heart Circ Physiol. 2015;309(11):1923–35. https://doi.org/10.1152/ajpheart.00284.2015.
Seemann F, Pahlm U, Steding-Ehrenborg K, Ostenfeld E, Erlinge D, Dubois-Rande J-L, Jensen SE, Atar D, Arheden H, Carlsson M, Heiberg E. Time-resolved tracking of the atrioventricular plane displacement in cardiovascular magnetic resonance (CMR) images. BMC Med Imag. 2017;17(1):19. https://doi.org/10.1186/s12880-017-0189-5.
Thavendiranathan P, Guetter C, da Silveira JS, Lu X, Scandling D, Xue H, Jolly M-P, Raman SV, Simonetti OP. Mitral annular velocity measurement with cardiac magnetic resonance imaging using a novel annular tracking algorithm: Validation against echocardiography. Magn Reson Imag. 2019;55:72–80. https://doi.org/10.1016/j.mri.2018.08.018.
Chen C, Qin C, Qiu H, Tarroni G, Duan J, Bai W, Rueckert D. Deep learning for cardiac image segmentation: A review. Front Cardiovasc Med. 2020;7:25. https://doi.org/10.3389/fcvm.2020.00025.
Hu C, Sinusas AJ, Huber S, Thorn S, Stacy MR, Mojibian H, Peters DC. T1-refBlochi: high resolution 3D post-contrast T1 myocardial mapping based on a single 3D late gadolinium enhancement volume, Bloch equations, and a reference T1. J Cardiovasc Magn Reson. 2017;19(1):63. https://doi.org/10.1186/s12968-017-0375-1.
Gonzales RA, Lamy J, Seemann F, Heiberg E, Onofrey JA, Peters DC. TVnet: Automated time-resolved tracking of the tricuspid valve plane in MRI long-axis cine images with a dual-stage deep learning pipeline. In: de Bruijne M, Cattin PC, Cotin S, Padoy N, Speidel S, Zheng Y, Essert C, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2021. Cham: Springer; 2021. p. 567–76. https://doi.org/10.1007/978-3-030-87231-1_55
Steding K, Engblom H, Buhre T, Carlsson M, Mosén H, Wohlfart B, Arheden H. Relation between cardiac dimensions and peak oxygen uptake. J Cardiovasc Magn Reson. 2010;12(1):8. https://doi.org/10.1186/1532-429X-12-8.
Steding-Ehrenborg K, Hedén B, Herbertsson P, Arheden H. A longitudinal study on cardiac effects of deconditioning and physical reconditioning using the anterior cruciate ligament injury as a model. Clin Physiol Funct Imag. 2013;33(6):423–30. https://doi.org/10.1111/cpf.12048.
Arvidsson PM, Töger J, Heiberg E, Carlsson M, Arheden H. Quantification of left and right atrial kinetic energy using four-dimensional intracardiac magnetic resonance imaging flow measurements. J Appl Physiol. 2013;114(10):1472–81. https://doi.org/10.1152/japplphysiol.00932.2012.
Erlinge D, Götberg M, Lang I, Holzer M, Noc M, Clemmensen P, Jensen U, Metzler B, James S, Bötker HE, Omerovic E, Engblom H, Carlsson M, Arheden H, Östlund O, Wallentin L, Harnek J, Olivecrona GK. Rapid endovascular catheter core cooling combined with cold saline as an adjunct to percutaneous coronary intervention for the treatment of acute myocardial infarction: The CHILL-MI trial: A randomized controlled study of the use of central venous catheter core cooling combined with cold saline as an adjunct to percutaneous coronary intervention for the treatment of acute myocardial infarction. J Am Coll Cardiol. 2014;63(18):1857–65. https://doi.org/10.1016/j.jacc.2013.12.027.
...Atar D, Arheden H, Berdeaux A, Bonnet J-L, Carlsson M, Clemmensen P, Cuvier V, Danchin N, Dubois-Randé J-L, Engblom H, Erlinge D, Firat H, Halvorsen S, Hansen HS, Hauke W, Heiberg E, Koul S, Larsen A-I, Le Corvoisier P, Nordrehaug JE, Paganelli F, Pruss RM, Rousseau H, Schaller S, Sonou G, Tuseth V, Veys J, Vicaut E, Jensen SE. Effect of intravenous TRO40303 as an adjunct to primary percutaneous coronary intervention for acute ST-elevation myocardial infarction: MITOCARE study results. Eur Heart J. 2014;36(2):112–9. https://doi.org/10.1093/eurheartj/ehu331.
Steding-Ehrenborg K, Boushel RC, Calbet JA, Åkeson P, Mortensen SP. Left ventricular atrioventricular plane displacement is preserved with lifelong endurance training and is the main determinant of maximal cardiac output. J Physiol. 2015;593(23):5157–66. https://doi.org/10.1113/JP271621.
Steding-Ehrenborg K, Arvidsson PM, Rydberg M, Carlsson M, Arheden H. Atrial and ventricular kinetic energy is higher in athletes compared to healthy controls and contributes to improve diastolic filling of the ventricles. J Cardiovasc Magn Reson. 2015;17(1):30. https://doi.org/10.1186/1532-429X-17-S1-P30.
Arvidsson PM, Kovács SJ, Töger J, Borgquist R, Heiberg E, Carlsson M, Arheden H. Vortex ring behavior provides the epigenetic blueprint for the human heart. Sci Rep. 2016;6(1):22021. https://doi.org/10.1038/srep22021.
Steding-Ehrenborg K, Arvidsson PM, Töger J, Rydberg M, Heiberg E, Carlsson M, Arheden H. Determinants of kinetic energy of blood flow in the four-chambered heart in athletes and sedentary controls. Am J Physiol Heart Circul Physiol. 2016;310(1):113–22. https://doi.org/10.1152/ajpheart.00544.2015.
Gyllenhammar T, Kanski M, Engblom H, Wuttge DM, Carlsson M, Hesselstrand R, Arheden H. Decreased global myocardial perfusion at adenosine stress as a potential new biomarker for microvascular disease in systemic sclerosis: a magnetic resonance study. BMC Cardiovasc Disorders. 2018;18(1):16. https://doi.org/10.1186/s12872-018-0756-x.
Stephensen SS, Ostenfeld E, Steding-Ehrenborg K, Thilén U, Heiberg E, Arheden H, Carlsson M. Alterations in ventricular pumping in patients with atrial septal defect at rest, during dobutamine stress and after defect closure. Clin Physiol Funct Imag. 2018;38(5):830–9. https://doi.org/10.1111/cpf.12491.
Bock J, Töger J, Bidhult S, Bloch KM, Arvidsson P, Kanski M, Arheden H, Testud F, Greiser A, Heiberg E, Carlsson M. Validation and reproducibility of cardiovascular 4D-flow MRI from two vendors using 2 × 2 parallel imaging acceleration in pulsatile flow phantom and in vivo with and without respiratory gating. Acta Radiologica. 2019;60(3):327–37. https://doi.org/10.1177/0284185118784981.
Töger J, Zahr MJ, Aristokleous N, Markenroth Bloch K, Carlsson M, Persson P-O. Blood flow imaging by optimal matching of computational fluid dynamics to 4D-flow data. Magn Reson Med. 2020;84(4):2231–45. https://doi.org/10.1002/mrm.28269.
Al-Mashat M, Jögi J, Carlsson M, Borgquist R, Ostenfeld E, Magnusson M, Bachus E, Rådegran G, Arheden H, Kanski M. Increased pulmonary blood volume variation in patients with heart failure compared to healthy controls: a noninvasive, quantitative measure of heart failure. J Appl Physiol. 2020;128(2):324–37. https://doi.org/10.1152/japplphysiol.00507.2019.
Heiberg E, Sjögren J, Ugander M, Carlsson M, Engblom H, Arheden H. Design and validation of segment - freely available software for cardiovascular image analysis. BMC Med Imag. 2010;10(1):1. https://doi.org/10.1186/1471-2342-10-1.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016;pp. 770–778. https://doi.org/10.1109/cvpr.2016.90
Deng J, Dong W, Socher R, Li L, Li K, Fei-Fei L. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, 2009;pp. 248–255. https://doi.org/10.1109/cvpr.2009.5206848
Onofrey JA, Casetti-Dinescu DI, Lauritzen AD, Sarkar S, Venkataraman R, Fan RE, Sonn GA, Sprenkle PC, Staib LH, Papademetris X. Generalizable multi-site training and testing of deep neural networks using image normalization. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 2019;pp. 348–351. https://doi.org/10.1109/isbi.2019.8759295
Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv e-prints, 2014;1412–6980.
Vigneault DM, Xie W, Ho CY, Bluemke DA, Noble JA. Ω-Net (Omega-Net): fully automatic, multi-view cardiac MR detection, orientation, and segmentation with deep neural networks. Med Image Analy. 2018;48:95–106. https://doi.org/10.1016/j.media.2018.05.008.
Dwyer M, Lyman C, Ferrari H, Bergsland N, Fuchs TA, Jakimovski D, Schweser F, Weinstock-Guttmann B, Benedict RHB, Riolo J, Silva D, Zivadinov R. DeepGRAI (Deep Gray Rating via Artificial Intelligence): Fast, feasible, and clinically relevant thalamic atrophy measurement on clinical quality T2-FLAIR MRI in multiple sclerosis. NeuroImage: Clinical 2021;30:102652. https://doi.org/10.1016/j.nicl.2021.102652
Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L, Summers RM. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imag. 2016;35(5):1170–81. https://doi.org/10.1109/tmi.2015.2482920.
Shin H, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imag. 2016;35(5):1285–98. https://doi.org/10.1109/tmi.2016.2528162.
Willemink MJ, Koszek WA, Hardell C, Wu J, Fleischmann D, Harvey H, Folio LR, Summers RM, Rubin DL, Lungren MP. Preparing medical imaging data for machine learning. Radiology. 2020;295(1):4–15. https://doi.org/10.1148/radiol.2020192224.
Tao Q, Yan W, Wang Y, Paiman EHM, Shamonin DP, Garg P, Plein S, Huang L, Xia L, Sramko M, Tintera J, de Roos A, Lamb HJ, van der Geest RJ. Deep learning-based method for fully automatic quantification of left ventricle function from cine MR images: A multivendor, multicenter study. Radiology. 2019;290(1):81–8. https://doi.org/10.1148/radiol.2018180513.
Hann E, Popescu IA, Zhang Q, Gonzales RA, Barutçu A, Neubauer S, Ferreira VM, Piechnik SK. Deep neural network ensemble for on-the-fly quality control-driven segmentation of cardiac MRI T1 mapping. Med Image Analy. 2021. https://doi.org/10.1016/j.media.2021.102029.
Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. https://doi.org/10.2307/1932409.
Acknowledgements
RAG acknowledges Magnus Caspersen, MSc for his guidance in deep learning, and DCP acknowledges James W. Goldfarb, PhD who realized the possibility of tracking valve insertion points with machine learning, many years ago. Authors would also like to acknowledge multiple researchers at Lund Cardiac MR Group for providing MV point delineation and collecting imaging data used in this study.
Funding
Open access funding provided by Lund University. The study have been funded by grants from the following providers: National Heart, Lung, and Blood Institute of the National Institute of Health (R01HL144706), Swedish Research Council, Knut and Alice Wallenberg Foundation, Region of Scania, Swedish Heart and Lung Foundation.
Author information
Authors and Affiliations
Contributions
RAG developed and implemented the algorithm, performed manual annotation in Yale dataset and data curation in both datasets, analyzed the results, drafted the manuscript, and helped conceive the study. FS and JL assisted in the design of the algorithm and the inter-observer analysis. HM included subjects for the Yale dataset. DA and DE initiated multi-vendor, multi-center clinical trials studies that were critical to enable data generalization. KSE included healthy volunteers and athlete subjects and supervised manual annotations in Lund dataset. HA, CH, JAO, DCP and EH contributed to the design of the algorithm and conceived the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All subjects in this study were part of previous research studies, and subjects provided written informed consent.
Consent for publication
All authors have read the final manuscript and approved for publication.
Competing interests
EH is the founder of the company Medviso AB that produces medical image analysis software. All other authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Mitral valve tracking in a Yale test sample by MVnet.
Additional file 2. Mitral valve tracking in a Lund test sample by MVnet.
Additional file 3.
Accuracy heatmap of inter-network variability. Each model (MVnet) trained on each training set was compared against other models on the mixed test set by the mean a Euclidean and b angular distance error, and the agreement with ICC in c MV displacement, d MAPSE, and e LV e'. ICC intra-class correlation coefficient, MV mitral valve, MAPSE mitral annular plane systolic excursion, LV left ventricle.
Additional file 4.
Clinical-metric agreement of LV s' (first row) and LV a' (second row) between an expert manual annotation (or observer 1) and a the automated method, and b annotation by a second group of observers, on a test set of 50 subjects. One observer from the Yale dataset and another from the Lund dataset annotated 25 subjects from each test set. In each scatter plot the black line denotes the identity line, whereas in each Bland-Altman plot, the red line denotes the mean difference (bias) and the two light dotted lines denote ± 1.96 standard deviations from the mean. LV left ventricle.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gonzales, R.A., Seemann, F., Lamy, J. et al. MVnet: automated time-resolved tracking of the mitral valve plane in CMR long-axis cine images with residual neural networks: a multi-center, multi-vendor study. J Cardiovasc Magn Reson 23, 137 (2021). https://doi.org/10.1186/s12968-021-00824-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12968-021-00824-2