Abstract
Background
Endometriosis is one of the most common gynaecological diseases, yet it lacks efficient biomarkers for early detection and unravels disease mechanisms. Proteomic profiling has revealed diverse patterns of protein changes in various clinical samples. Integrating and systematically analysing proteomics data can facilitate the development of biomarkers, expediting diagnosis and providing insights for potential clinical and therapeutic applications. Hence, this systematic review and meta-analysis aimed to explore potential non-invasive diagnostic biomarkers in various biological samples and therapeutic targets for endometriosis.
Methods
Online databases, including Scopus, PubMed, Web of Science, MEDLINE, Embase via Ovid, and Google Scholar, were searched using MeSH terms. Two independent authors screened the articles, extracted the data, and assessed the methodological quality of the included studies. GO and KEGG analyses were performed to identify the pathways that were significantly enriched. Protein‑protein interaction and hub gene selection analyses were also conducted to identify biomarker networks for endometriosis.
Results
Twenty-six observational studies with a total of 2,486 participants were included. A total of 644 differentially expressed proteins (180 upregulated and 464 downregulated) were identified from 9 studies. Proteins in peripheral blood exhibited a sensitivity and specificity of 38-100% and 59-99%, respectively, for detecting endometriosis, while proteins in urine had a sensitivity of 58-91% and specificity of 76-93%. Alpha-1-antitrypsin, albumin, and vitamin D binding proteins were significantly DEPs in both serum and urine. Complement C3 is commonly expressed in serum, menstrual blood, and cervical mucus. Additionally, S100-A8 is commonly expressed in both menstrual blood and cervical mucus. Haptoglobin is commonly detected in both serum and plasma, whereas cathepsin G is found in urine, serum, and plasma. GO and KEGG enrichment analyses revealed that proteoglycans in cancer pathways, which regulate cell-to-cell interactions, modulate the extracellular matrix, and promote the proliferation and invasion of endometrial cells, are commonly enriched in serum and urine.
Conclusion
This comprehensive study revealed potential proteomes that were significantly differentially expressed in women with endometriosis utilizing various non-invasive clinical samples. Exploring common differentially expressed proteins in various biological samples provides insights into the diagnosis and pathophysiology of endometriosis, as well as potential clinical and therapeutic applications.
Graphical abstract

Similar content being viewed by others
Background
Endometriosis is characterized by the development of endometrium-like tissue and/or stroma outside the endometrium and myometrium [1, 2]. It is a chronic inflammatory disease that affects more than 170 million women worldwide, predominantly women of reproductive age, with a wide range of clinical symptoms, including dysmenorrhea, dyspareunia, dyschezia, dysuria, chronic pelvic pain, and infertility, affecting women’s health from the time of menarche to menopause, regardless of their ethnicity or social status [1, 3].
In clinical settings, the gold standard diagnostic method for confirming endometriosis is laparoscopy, a minimally invasive surgical procedure that involves inserting an imaging tube through a small incision in the abdomen [4]. Although laparoscopy is effective and the gold standard, it has potential complications, requires general anaesthesia, and demands advanced surgical skills [5,6,7]. Moreover, it is not always available or accessible, particularly in low- and middle-income countries where healthcare facilities and resources are lacking [5]. Ultrasound is the first-line non-invasive diagnostic method for detecting endometriosis [8]. It has been widely used to enhance the diagnosis and identification of endometriomas and nodules in adjacent structures of the pelvis but lacks both sensitivity and specificity for ruling out peritoneal endometriosis, endometriosis-associated adhesions, and deep infiltrating endometriosis [9, 10]. Imaging techniques such as transvaginal ultrasound (TVS), transrectal ultrasound (TRS), and magnetic resonance imaging (MRI) can bridge the gap between clinical and surgical diagnosis by providing a non-invasive visual diagnosis that can be achieved more quickly, safely, and accessibly than surgery. However, different studies have reported wide variation in diagnostic accuracy between MRI and TVS, mainly due to the variability of techniques, examiners’ experience, and anatomic locations of the lesions/subtypes of the disease [11]. Given these challenges, non-invasive diagnostic approaches for endometriosis are urgently needed.
While various non-invasive diagnostic modalities involving blood, cervicovaginal fluid, and urine have been proposed, a definitive diagnostic biomarker for endometriosis remains elusive. Despite extensive research into blood and urine tests and the investigation of altered levels of cytokines, angiogenic factors, and growth factors, none of these biomarkers have been used to conclusively diagnose endometriosis [12,13,14,15]. In addition, numerous studies have demonstrated that nanoparticles, which are materials with dimensions smaller than 100 nanometers, hold promise for improving diagnostic and imaging techniques for non-invasive detection, understanding target signalling pathways, and identifying therapeutic options for diverse diseases [16,17,18,19]. Notably, nanoparticles can serve as carriers for transporting anti-inflammatory, antioxidant, anti-angiogenic, or immunomodulatory molecules to specific locations [20,21,22,23], owing to their low toxicity, high stability, and capacity for conjugating with various biomolecules [21, 24, 25]. Moreover, nanotechnology may offer a promising non-invasive diagnostic method for detecting endometriosis by identifying specific biomarkers, such as proteins or genetic materials [26]. Although studies have shown that CA 19 − 9 and CA-125 have been detected in blood using immunochemical sensing [27, 28], the recognition of iron oxide nanoparticles as contrast agents for magnetic resonance imaging [26, 29], and the investigation of gold nanorods and carbon nanotubes as photoacoustic imaging agents for visualizing endometriosis lesions in vivo [26, 30]. However, it is important to note that none of the biomarkers/methods have been clinically proven biomarkers for endometriosis detection. Among all techniques, proteomic approaches are essential for identifying biomarkers by characterizing the protein content of biological samples [31]. These approaches enable proteome profiling, comparative expression analysis of proteins in various biological samples, identification of posttranslational modifications, and identification of protein–protein interactions. Notably, proteomic analysis is invaluable because proteins, unlike DNA or RNA, directly mediate cellular functions and disease mechanisms [32, 33]. Mass spectrometry (MS) proteomic methods have appeared to be powerful platforms for discovering novel and potential diagnostic and prognostic biomarkers for various diseases. MS-based approaches are substantially helpful for consistently identifying proteins with high diagnostic accuracy for endometriosis [34]. Furthermore, proteomics studies offer functional insights into expressed proteins and significantly enriched pathways, providing valuable information for understanding the pathogenesis of this disease.
Our hypothesis is that biomarkers of endometriosis commonly found in various biological samples may have substantial significance and have a direct impact on the development and progression of endometriosis. Therefore, our aim is to gain a thorough understanding of the diagnosis, pathogenesis, and possible therapeutic approaches for endometriosis utilizing diverse clinical samples, which could ultimately result in improved patient outcomes and quality of care. Hence, this systematic review aims to assess the utility of proteomic (MS-targeted) analysis for biomarker discovery and navigate the pathogenesis of endometriosis development. Additionally, this study explored the sensitivity and specificity of expressed proteins as promising biomarkers for detecting endometriosis. Moreover, this study involved mass spectrometry-based diagnostic testing for endometriosis and a comprehensive understanding of the pathogenesis of endometriosis in various non-invasive biological samples, including peripheral blood, cervical mucus, menstrual blood, and urine. Remarkably, this study examined commonly enriched pathways associated with disease conditions to better understand the mechanism of disease development.
Methods
Protocol registration
Following the PRISMA 2020 checklist, we conducted a systematic review and registered the protocol with PROSPERO (registration ID: CRD42023397217).
Study search strategy
Searches were performed in the following databases: PubMed, EMBASE through OVID, Google Scholar, Scopus, and Web of Science. The following terms were used in the search strategy, with alternatives as shown using Boolean operators: “mass spectrometry” AND (“diagnostic” OR “test”) AND (“endometriosis” OR “endometrioma”) & (‘’proteomics’’ OR’’ proteome’’ AND (‘’endometriosis’’ OR’’ endometrioma”).
In addition, manual searches were performed for the reference lists of all studies identified by the search strategy described above. Web sources and databases were searched for published articles and preprint research papers written in the English language up to January 31, 2024.
Study participants
The participants in the study were reproductive-aged women who underwent laparoscopy or abdominal surgery for one of the following reasons: pelvic pain, infertility, dysmenorrhea, abnormal pelvic examination, or a combination of the aforementioned conditions, an ovarian mass regardless of symptoms, or other pelvic pathologies. Only confirmed cases with laparoscopy and/or histology data were included in the review after surgery, while women with confirmed benign pelvic pathologies, such as uterine fibroids, ovarian cysts, unexplained infertility, and fertile healthy women were considered as controls.
Study selection
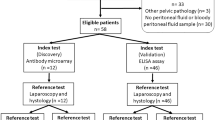
From the initial 2,273 retrieved articles, we included 22 case-control, 2 cross-sectional, and 2 prospective cohort studies that met our eligibility criteria. Laparoscopy or laparotomy with or without histological confirmation and mass spectrometry techniques were used as reference standards and index tests, respectively.
Inclusion criteria
In this study, women with a confirmed diagnosis of endometriosis, either combined with one phenotype (I) ovarian endometriosis, (II) deep pelvic infiltrating endometriosis (DIE), and (III) peritoneal endometriosis, were enrolled as cases, whereas women with benign uterine conditions such as uterine fibroids and ovarian cysts and healthy women (self-declaration) were enrolled as controls. All observational studies, such as cohort, cross-sectional, and case-control studies, that were published exclusively in the English language were considered for inclusion.
Exclusion criteria
Endometriosis with other coincidental pelvic pathologies, such as pelvic malignancy, adenomyosis, polycystic ovarian syndrome (PCOS), and pelvic inflammatory disease (PID), studies conducted with approaches other than mass spectrometry-based series, proteomics studies with invasive sources of samples, such as peritoneal fluid, endometrial biopsy, follicular fluid, and endometrial fluid, and studies reporting proteins with other index tests, such as enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR/qPCR), and western blot, were excluded from the study. Additionally, case reports or series, articles without full text and abstracts, duplicated studies, anonymous reports, editorial reports, reviews, perspectives, and book sections or chapters were also excluded.
Data extraction
The authors’ names, year of study, country, diagnostic criteria for endometriosis, type of sample, protein alterations, menstrual phase, proteomics platform, sensitivity, and specificity of biomarkers with a molecular weight of m/z were extracted from each article (Table 1). In addition, for the bioinformatics analysis, the protein ID (UniProt), protein accession, and fold change (up- and downregulated) were extracted. Moreover, the protein lists from the 8 articles were extracted, including the identification codes and the level of regulation (up/downregulated). The UniProt website (https://www.uniprot.org/) was used to standardize the protein identification codes. Subsequently, a comparison was conducted on the significantly differentially expressed proteins extracted from the 8 papers to identify consistent proteins. Studies reporting the p value (p < 0.05) and fold change (FC) of differentially expressed proteins were included in the meta-analysis.
Risk of bias and applicability
Two authors (GGA & BKA) conducted independent assessments of risks associated with bias and applicability using the Diagnostic Precision Study Quality Assessment Tool (QUADAS-2) for the studies included in the diagnostic accuracy review [35]. Conflicts between the two authors were evaluated and reviewed by a third author (LW). Patient selection, index test, reference standard, and flow and timing were the four domains used to evaluate the risk of bias, whereas patient selection, index test, and reference standard were the domains employed to assess the applicability of each article. The distribution of risk-of-bias and applicability judgments within each bias domain was assessed (Figure S1).
Identification and enrichment of DEPs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to elucidate the biological characteristics of the overlapping DEGs via the online tool database for annotation, visualization, and integrated discovery (DAVID) (https://david.ncifcrf.gov/). GO annotation and KEGG pathway analyses were performed with Metascape (https://metascape.org/). Furthermore, a science and research online plot (SRplot) (https://www.bioinformatics.com.cn/en) was used to present the findings. GO and KEGG analyses were performed for each clinical sample separately, such as peripheral blood (serum, plasma), urine, and menstrual blood. DEPs from the supernatant and mesenchymal stem cells derived from menstrual blood were combined and analysed as menstrual blood-expressed proteins. For each given gene list, pathway and process enrichment analyses were carried out with KEGG and GO pathway analyses. Metascape (https://metascape.org/) default parameters: terms with a p value < 0.05, a minimum count of 3, and an enrichment factor > 1.5 were deemed significant. Moreover, p values are calculated based on the cumulative hypergeometric distribution, and q-values are calculated using the Benjamini‒Hochberg procedure to account for multiple tests [36].
Protein‒protein interaction (PPI) network construction and analysis
The PPI network was constructed with the STRING (https://string-db.org/) database with a threshold of a combined score > 0.4, and the interaction networks were visualized with Cytoscape (version 3.10.1). In addition, the molecular complex detection (MCODE) plug-in was used to screen strongly interconnected modules in the PPI network with default parameters (degree cut-off = 2, node score cut-off = 0.2, and K-score = 2).
Hub gene selection and analyses
The Cyto-Hubba plug-in in Cytoscape (version 3.10.1) was used to select hub genes in the PPI network. Based on the evidence in the literature, we selected five of the 12 algorithms in the cyto-Hubba plug-in and took the intersection of the five parameters (degree, edge percolated component, maximum neighborhood component, maximal clique centrality, and eccentricity) to determine the hub genes in each biological sample.
Results
Study characteristics
A total of 2,273 articles were identified from the online databases with the search strategy. After removing 351 duplicate results, 1922 articles remained. Moreover, 1851 articles were excluded after reviewing the title and abstract, and 71 articles met the eligibility criteria for full-text review and further consideration. Finally, 26 of the 71 identified articles met the eligibility criteria. All selected studies were performed in Asian, American, and European countries (9 in China, 1 in India, 1 in Japan, 3 in the USA, 2 in South Korea, 1 in Belgium, 1 in Germany, 1 in Austria, 2 in Italy, 1 in Australia, 2 in the UK, 1 in Brazil, and 1 in Croatia). Platforms for proteomics included surface-enhanced laser desorption/ionization-time of flight mass spectrometry (SELDI-TOF-MS) (8 studies), SOMA scanning (1 study), electrospray ionization quadrupole time-of-flight mass spectrometry (ESI-Q-TOF-MS) (1 study), liquid chromatography‒mass spectrometry (LC‒MS/MS) (9 studies), and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF/MS) (7 studies). The biological samples included in this study were peripheral blood (15 studies), urine (8 studies), cervical mucus (1 study), and menstrual blood (2 studies). A PRISMA flow chart that depicts each step is shown in Fig. 1. The studies analysed in this review were all conducted from 2007 to 2023, and a total of 2,486 women were enrolled.
Diagnostic accuracy of proteins
Various proteomic techniques have been used to investigate potential biomarkers for detecting endometriosis. Peripheral blood (serum and plasma) protein biomarker analysis has a sensitivity of 38–100% and a specificity of 59–99% for detecting endometriosis (Table 2). Additionally, urine proteomic profiling revealed that single and/or combined proteins could detect endometriosis with a sensitivity ranging from 58 to 91% and a specificity ranging from 76 to 93% (Table 2).
Common DEPs in endometriosis
In endometriosis, different proteins are expressed in various biological samples. Peroxiredoxin-6, angiopoietin-related protein, heterogeneous nuclear ribonucleoproteins, peroxiredoxin-1, leucine-rich alpha-2-glycoprotein, alpha-2-macroglobulin, apolipoprotein L1 and haptoglobin are commonly expressed proteins in plasma and serum samples. Alpha-1-antitrypsin, alpha-enolase, albumin, and vitamin D-binding protein are commonly expressed in both urine and serum, whereas S100-A8 and complement proteins are expressed in cervical mucus and menstrual blood as well as serum. Additionally, dynamin-1-like protein, rho GTPase-activating protein 6, rho GTPase-activating protein 18, zinc finger protein 185, FYN-binding protein 1, rho GTPase-activating protein 45, neurosecretory protein VGF, cartilage oligomeric matrix protein, stromal interaction molecule 1, polymeric immunoglobulin receptor, adipogenesis regulatory factor, complement C3, serum amyloid A-1 protein, fibrinogen gamma chain and ATP-dependent RNA helicase A are differentially expressed proteins in both serum and menstrual blood (Fig. 2 and Table S1).
The distribution of DEPs (overexpressed) in endometriosis patients in different clinical samples supplemented with Table S1: List of differentially expressed proteins
GO, KEGG, and PPI analyses of the DEPs in women with endometriosis
A total of 644 DEPs (180 upregulated and 464 downregulated) were identified from 9 studies in different clinical samples, such as peripheral blood (serum, plasma), menstrual blood, cervical mucus, and urine. Among these studies, 8 met the eligibility requirements for meta-analysis, and the remaining cervical mucus clinical samples were comprehensively reviewed and described (Fig. 3).
Plasma
The DEPs from plasma samples were analysed using GO terms that were categorized into molecular functions, cellular components, and biological processes. The molecular functions of the DEPs were primarily enriched in signalling receptor activator activity, signalling receptor regulator activity, and kinase activity. The GO terms in the cellular component category were mainly related to the collagen-containing extracellular matrix, the external secretory granule lumen, and the extracellular matrix. The biological process GO terms were primarily involved in the regulation of cell activation, regulation of leukocyte activation, and regulation of lymphocyte activation (Fig. 4 & Fig. S6). The enriched GO networks are also illustrated in Fig. S2.
We conducted KEGG pathway enrichment analysis of DEPs from plasma samples to explore DEP-related gene pathways in endometriosis. Nitrogen metabolism pathways, the phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) pathway, and microRNAs in cancer pathways were the most significant (Fig. 4 & Fig. S7). In general, GO and KEGG analyses revealed that cell proliferation, adhesion, migration, and inflammation are involved in the pathophysiology of endometriosis.
PPI network analysis was performed for the 69 DEPs using the STRING database. After removing proteins without standard symbols, a total of 68 nodes and 121 edges were obtained that represented the interaction network with a p value of 1.98e-10. The top five hub genes identified using the cyto-Hubba plugin included casein kinase II subunit alpha (CSNK2A1, CSNK2A2), mammalian topoisomerase 1 (TOP1), cAMP-dependent protein kinase catalytic subunit alpha (PRKACA) and RNA-binding protein 39 (RBM39) (Fig. 2). The MCODE plugin distinguished two cluster networks, and all the top five hub genes, CSNK2A2, CSNK2A1, TOP1, PRKACA and RBM39, were included in the cluster with the highest score.
Serum
Category-based GO analysis of the DEPs from serum samples was performed. The cellular component of the DEPs was predominantly enriched in collagen-containing extracellular matrix binding, extracellular matrix, and secretory vesicle lumen. The molecular function category was mainly involved in cell adhesion molecule binding, kinase binding, and actin binding (Fig. 4 & Fig S6). Actin filament organization, supramolecular fiber organization, and regulation of body fluid levels are predominantly involved in biological processes. The enriched GO term networks are also illustrated in Fig S3.
KEGG enrichment pathway analysis was carried out on serum samples to elucidate the pathogenesis of endometriosis. The top ten enriched pathways are illustrated in Fig. 4 and Fig. S7. The complement and coagulation cascades, platelet activation, neutrophil extracellular trap formation, and tight junction pathways were the most enriched KEGG pathways.
PPI network analysis of the 428 DEPs in serum was performed using the STRING database. A total of 396 nodes and 3186 edges associated with the PPI network were identified after removing proteins with no symbol name (PPI enrichment p value: < 1.0e-16). The five top hub genes were identified using the cytoHubba plugin and included albumin (ALB), actin, cytoplasmic 1 (ACTB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), fibronectin (FN1), and apolipoprotein A-I (APOA1) (Fig. 2). The MCODE plugin distinguished two cluster networks, and all the top five hub genes, ALB, ACTB, GAPDH, FN1, and APOA1, were included in the cluster with the highest score.
Menstrual blood
GO analysis demonstrated that DEPs derived from menstrual blood are involved in the pathophysiology of endometriosis. In addition, the GO analysis results were categorized into three components, i.e., molecular functions, cellular components, and biological processes. The molecular functions of the DEPs were mainly enriched in protease binding, receptor‒ligand activity, and fatty acid binding. The GO terms in the cellular component category were mainly involved in the vesicle lumen, secretory granule lumen and cytoplasmic vesicle lumen (Fig. 4 & Fig. S6). Granulocyte migration, granulocyte chemotaxis, and leukocyte chemotaxis are the main biological processes involved. The enriched GO networks are also shown in Fig. S4.
The enriched KEGG pathways of DEPs from menstrual blood samples were used to further investigate DEP-related gene pathways. The top ten enriched pathways are illustrated in Fig. 4 and Fig. S7. IL-17 signalling pathway, complement and coagulation cascades, cytokine‒cytokine receptor interaction, and TNF signalling pathway. In conclusion, the GO and KEGG enrichment pathway analyses revealed that angiogenesis, cell proliferation, differentiation, and the induction of inflammation are highly important for the pathogenesis of endometriosis.
The STRING database was used for the PPI network analysis of 110 DEPs. After identifying proteins with no symbol name, there were 89 nodes and 134 edges associated with the PPI network (p value < 1.0e-16).
The top five hub genes identified using the cyto-Hubba plugin included protein S100 calcium-binding protein A9 (S100-A9), C-X-C motif chemokine ligand 1 (CXCL1), interleukin-1 receptor antagonist protein (IL1RN), cystatin-A (CSTA), and protein S100-A8 (Fig. 2). The MCODE plugin illustrated three cluster networks (cluster one: 7 nodes (desmoglein 1 & 3 (DSG1&DSG3), small proline-rich protein 3 (SPRR3), CSTA, small proline-rich protein 1B (SPRR1B), ajuba LIM protein (JUB) and serpin Family B Member 13 (SERPINB13), 19 edges; cluster two: 7 nodes (S100-A8, S100-A9, myeloperoxidase (MPO), IL1RN, C-X-C motif chemokine ligand 1 & 5 (CXCL1, CXCL5); and growth differentiation factor 15 (GDF15), 14 edges; and cluster three: 3 nodes (haptoglobin (HP), cyclic adenosine 3′,5′-monophosphate (CAMP) and resistin (RETN) and 3 edges). The top five hub genes, S100A9, IL1RN, CSTA, S100A8, and CXCL1, were included in the cluster with the highest score.
Urine
The three categories of GO term analysis, i.e., molecular functions, cellular components, and biological processes, of the DEPs from urine samples were notably involved in the pathophysiology of endometriosis. The molecular functions of the DEPs were mainly enriched in collagen binding, cytokine binding, and transforming growth factor binding. The GO terms in the cellular components category were mainly involved in the collagen-containing extracellular matrix, secretory vesicle lumen, and extracellular matrix (Fig. 4 & Fig. S6). Cell‒cell adhesion, plasminogen activity regulation, and body fluid level regulation are the main biological processes involved. The enriched GO networks are also illustrated in Fig S5.
The KEGG pathway enrichment of DEPs from urine samples revealed the DEP-related gene pathways that are involved in the mechanism of endometriosis pathogenesis. ECM receptor interactions and microRNAs in cancer pathways were the pathways most significantly associated with endometriosis development (Fig. 4 & Fig. S7). Generally, GO and KEGG analyses revealed that cell growth and invasion, adhesion, and angiogenesis were implicated in the pathophysiology of endometriosis.
PPI network analysis of the 22 DEPs was performed using the STRING database, which revealed 22 nodes and 39 edges associated with the PPI network (p value: 9.7e-14). The top five hub genes identified using the cyto-Hubba plugin included thrombospondin-1 (THBS1), albumin (ALB), CD44 antigen (CD44), annexin A2 (ANXA2), and (LUM) (Fig. 2). The MCODE plugin distinguished two cluster networks. In cluster one, CD44, alkaline phosphatase (ALP), zinc-alpha-2-glycoprotein (AZGP1), alpha-1-antitrypsin (SERPINA1), ANAX2, and enolase 1 (ENO1) were the most sub connected proteins, whereas transforming growth factor beta receptor 2 (TGFBR2), endoglin (ENG), THBS1 and LUM were the most highly connected subnetworks in cluster two.
Discussion
This is a comprehensive systematic review and meta-analysis of proteomics data to explore common pathways and non-invasive diagnostic biomarkers for detecting endometriosis. Proteomic platforms offer an extraordinary opportunity to overcome the challenges associated with endometriosis by providing valuable insights into the mechanisms underlying the disease and identifying potential markers for diagnosis and therapeutic targeting. Hence, this study focused on recent improvements in proteomics technology aimed at identifying potential non-invasive diagnostic biomarkers and establishing mechanistic pathways to understand the pathogenesis of endometriosis.
Alteration of proteins in endometriosis
This study investigated DEPs in peripheral blood, cervical mucus, menstrual blood, and urine from women with endometriosis. Although many proteins are altered in women with endometriosis, this review illustrates the common DEPs in diverse biological samples from women with endometriosis. DEPs commonly found in multiple biological samples, including vitamin D binding protein (VDBP), haptoglobin, S100-A8, cathepsin G, and complement component 3, are discussed.
VDBP is one of the most common proteins whose expression is altered in women with endometriosis. A line of evidence has shown that the expression of VDBP is substantially increased in the urine [37] and serum [38] of women with endometriosis compared to women without endometriosis. Similarly, the expression level of VDBP is markedly higher in endometrial tissue [39] but lower in peritoneal fluid [40] in women with endometriosis. Although studies have shown that VDBP may be implicated in the pathogenesis of endometriosis because of its chemotactic characteristics and ability to attract immune cells [39, 40], inconsistent patterns of VDBP expression have been observed across studies. The potential reasons for discrepancies may be observed in various studies, attributing them to differences in biological specimens, protein extraction procedures, centrifugal forces, and analysis platforms. Regarding the abundance of VDBP, studies have described diverse techniques for sample handling and analysis, such as 2DE-gel electrophoresis with LC‒MS/MS [37, 38] and ELISA [41, 42]. These disparities highlight the potential influence of methodological applications, as evidenced by (1) the superior sensitivity of LC‒MS/MS compared to ELISA, (2) the possibility of cell loss in the supernatant, affecting the abundance and concentration of proteins when employing low centrifugation force or short processing time, and (3) the superior sensitivity and ability of ELISA to detect very small amounts of target proteins compared to 2DE-gel electrophoresis [43, 44]. These perspectives highlight the clinical utility of LC‒MS/MS, which is a standard and high-throughput proteomics technology with a lesser tendency for bias or interference, as well as greater quantitative agreement among laboratories and biological samples [45, 46]. Given the wide range of variation within biological samples that does not adequately explore protein alterations across the severity and phenotype of endometriosis, conducting further large-scale multi-omics studies would be helpful to elucidate the association between VDBP and the underlying mechanism of endometriosis.
The expression level of haptoglobin decreased in the plasma and serum of women with endometriosis [47]. However, this finding contradicts the findings of Wölfler et al., who demonstrated that the alteration of haptoglobin is significantly increased in the peritoneal fluid of patients with ovarian and peritoneal endometriosis [48]. The potential variation may be due to the diverse phenotypes of endometriosis, including ovarian, peritoneal, and deep endometriotic lesions, as well as the timing of sample collection. The upregulation of estrogen and the estrogen receptor on macrophages in the peritoneal cavity generates an abnormal immune microenvironment, potentially resulting in increased haptoglobin production [49]. In addition to the phenotype of endometriosis, the depletion of proteins should also be considered for the variations that ensue. Some studies depleted the most abundant proteins, such as albumin and globulin, to detect low-abundance proteins, which may be putative disease biomarkers in biological samples [47], whereas other studies did not mention the depletion process during protein extraction and identification [50,51,52]. Therefore, protein depletion can affect the haptoglobin concentration during protein extraction via different mechanisms, including reduced solubility, altered protein‒ligand interactions, and competitive binding [53,54,55]. The proteomics analysis platform is also another confounding factor. The two common analysis platforms are mass spectrometry and enzyme-linked immunoassay. Both techniques are used to detect the concentration and expression of proteins. However, compared with ELISA, mass spectrometry (MS)-based proteomics analysis [47, 50] provides high accuracy, resolution, reproducibility, and sensitivity in identifying and quantifying proteins in a complex mixture, often not allowing differentiation between the peptide and its derivatives or degradation fragments [49, 56].
This study similarly demonstrated that the protein S100-A8 is markedly reduced in the cervical mucus of women with endometriosis. This finding supports the findings of a study conducted in France, which identified S100-A8 as a promising endometrial diagnostic marker for both the proliferative and secretory phases [57]. Additionally, another study showed that S100A8 is predominant in the peritoneal fluid of women with early-stage deep endometriosis [51]. In addition, the presence of higher levels of S100A8 in the peritoneal fluid of women with endometriosis suggests its potential contribution to the development and formation of lesions within the peritoneal cavity through inflammatory pathways by activating neutrophils [58, 59].
This study also revealed that cathepsin G is a common DEP in the urine, serum, and plasma of women with endometriosis. This finding supports the findings of a study conducted in Poland, which revealed that cathepsin G is significantly elevated in the endometrial tissue of women with endometriosis and may play a role in disease development and progression [60]. Several lines of evidence have demonstrated that cathepsin G plays an essential role in the pathogenesis of endometriosis by promoting extracellular matrix degradation and invasion [61], activating collagen production [61], and stimulating the inflammatory process [62], which facilitates the implantation and growth of endometrial tissue outside the uterus.
This comprehensive study also showed that complement C3 levels are significantly higher in women with endometriosis than in those without endometriosis. Similarly, it has been reported that the abundance of C3 is significantly higher in peritoneal fluid [63] and endometrial tissue [64, 65] in women with endometriosis. The involvement of complement C3, as expressed by ectopic endometrial tissue, in the formation of endometriotic lesions is mediated by mast cell activation. Additionally, it may be generated locally by ectopic endometrial tissue and can promote the engraftment of endometriotic cysts [65, 66]. Moreover, cyto-hub gene analysis revealed that CSNK2A1, CSNK2A2, TOP1, PRKACA, RBM39 (plasma), ALB, ACTB, GAPDH, FN1, APOA1 (serum), S100-A9, CXCL1, IL1RN, CSTA, S100-A8 (menstrual blood) and THBS1, ALB, CD44, ANXA2, and LUM (urine) were the top 5 proteins expressed in women with endometriosis. Among all the proteins, ALB is commonly expressed in both serum and urine. These disparities were also revealed by a study conducted by Donal S et al., who reported that the percentages of proteins in venous blood, menstrual blood, and vaginal fluid were 61%, 36%, and 35%, respectively. These body fluid-derived proteins could contribute to augmenting the diagnosis of endometriosis combined with imaging techniques and physical examinations. Nevertheless, to enhance the diagnostic accuracy of non-invasive biological sample-derived proteins, further comprehensive functional and validation multi-omics studies with large sample sizes are needed.
GO analysis revealed that the modulation of molecular, functional, and cellular processes contributes to the pathophysiology of endometriosis through the activation of the collagen-containing extracellular matrix, extracellular matrix, secretory granule lumen, and others [67]. These GO terms play a role in cell migration, adhesion, angiogenesis, immune response, lymphocyte activation, tissue survival, and facilitating the implantation and potential growth of ectopic endometrial lesions [13, 68,69,70,71].
KEGG enrichment analysis revealed that nitrogen metabolism [72], PI3K-Akt [73], platelet activation [74], the NOD-like receptor signalling pathway [75], ECM-receptor interactions [76], cytokine‒cytokine receptor interactions [76], IL-17 signalling [77], complement and coagulation cascades [78], TNF signalling and proteoglycans in cancer [79] have been implicated in the pathogenesis of endometriosis. These pathways play a significant role in the cellular growth and survival of endometriotic lesions [80,81,82]. The ECM pathway plays a key role in cell migration, adhesion, and tissue remodelling through the modulation of matrix metalloproteinases that interact with various growth factors and proinflammatory cytokines, such as transforming growth factor-beta (TGF-β), interleukin-1 (IL-1), and tumor necrosis factor-alpha (TNF-α) [83,84,85].
The NOD-like receptor pathway is an important signalling pathway that is involved in the pathogenesis of endometriosis [86]. This pathway encompasses the family pyrin domain containing 3 (NLRP3), an intracellular receptor that initiates the release of proinflammatory cytokines such as interleukin-1β (IL-1β) upon the activation of NLRP3. Abnormal activation of the NLRP3 inflammasome has been observed within ectopic endometrial lesions, peritoneal fluid, and the eutopic endometrium of women with endometriosis. This dysregulated activation significantly contributes to persistent inflammation and accompanying pain related to the condition [86,87,88,89]. Cytokine‒cytokine receptor interactions and the IL-17 signalling pathway have been implicated in the pathogenesis of endometriosis. IL-17 has been shown to promote the production of other proinflammatory cytokines, such as IL-1α and IL-1β, involved in the pathogenesis of endometriosis [77]. Additionally, an interaction between the complement system and coagulation system might contribute to the pathophysiology of endometriosis following the monthly shedding of endometrial tissues, triggering complement activation resulting from the activation of the microenvironment in women diagnosed with endometriosis [90].
Proteoglycans involved in cancer pathways are commonly enriched in both the serum and urine of women with endometriosis. Proteoglycans are complex molecules that are secreted by cancer cells and stromal cells and are composed of glycosaminoglycan (GAG) chains [91]. The literature has shown that proteoglycans play a significant role in regulating cell-to-cell and cell-to-matrix interactions, releasing growth factors and cytokines that can promote cell proliferation and invasion [92]. Hence, the trapping and release of angiogenic factors and cytokines that trigger proliferation and invasion are implicated in the pathophysiology of endometriosis.
Overall, this proteomics study provides insights into the expression of common and distinct proteins that are expressed in women with endometriosis. Given the different conditions of the study participants, the phenotype and severity of endometriosis, sample handling, and processing methods, proteomic platforms, and different menstrual cycles, we recommend the use of an integrated multi-OMICS study in which all non-invasive biological samples from the same patients are adjusted for confounders to enhance the mechanism of disease development and provide an opportunity to identify novel diagnostic and therapeutic targets for endometriosis (Fig. 5).
Strengths and limitations
This is a comprehensive systematic review and meta-analysis to explore the applicability of the proteomics approach to discover novel diagnostic biomarkers and unravel therapeutic targets from non-invasive biological samples. Additionally, this study serves as an input for further multi-OMICS studies to uncover and establish novel diagnostic and therapeutic targets in endometriosis. There are some limitations in our study. First, there is a lack of sufficient studies on the overall diagnostic accuracy of individual or combined proteins based on the expression molecular weight of proteins/peptides in different phases of the menstrual cycle. Although the literature has shown protein expression in endometriosis during different phases of the human menstrual cycle, the difference in protein expression between the proliferative and secretory phases remains controversial. Therefore, further evidence is required to explore the diagnostic accuracy of protein biomarkers concerning the m/z ratio in different phases of the menstrual cycle. Second, the lack of available raw data and/or full protein lists allowed us to focus only on the differentially expressed protein lists, which could affect the conclusions of the findings. Additionally, the lack of studies did not allow us to look at the differentially expressed proteins across the stages (early vs. advanced, subtypes of endometriosis (ovarian, peritoneal & deep infiltrating) and menstrual cycles (secretary, proliferative and menstrual phases).
Conclusion
In summary, this comprehensive meta-analysis of differentially expressed proteins from non-invasive clinical samples highlights the pathophysiology of endometriosis with GO and enriched KEGG pathways. Moreover, proteomics holds promise for the discovery of peripheral blood, menstrual blood, cervical mucus, and urine-based biomarkers for endometriosis. Various upregulated and downregulated proteins have been identified, suggesting their potential utility as promising non-invasive biomarkers for endometriosis detection and disease development mechanisms.
Furthermore, this review explored how the expression of different proteins and pathways in multiple clinical samples from non-invasive sources can be used to elucidate the pathophysiology of endometriosis. Finally, our findings provide new knowledge that will be helpful in understanding the pathophysiology of endometriosis, and future integrated studies involving peripheral blood, menstrual blood, and urine samples are needed. The identified proteins and pathways not only expand our understanding of the disease but also offer promising targets for future research. Furthermore, validation of these findings, exploration of hub genes for diagnostic accuracy, and further research across a wider range of samples and endometriosis types are key to revealing new options for non-invasive diagnosis and helping to explore more effective potential treatment options. Moreover, further research is needed to validate these findings and potentially help to improve the diagnosis, enhance pathophysiology, and offer hints for potential treatments for endometriosis.
Data availability
The data underlying this article are available upon the request of the corresponding authors.
References
Delanerolle G, Ramakrishnan R, Hapangama D, Zeng Y, Shetty A, Elneil S, et al. A systematic review and meta-analysis of the endometriosis and mental-health sequelae; the ELEMI project. Women’s Health. 2021;17:17455065211019717.
Becker CM, Bokor A, Heikinheimo O, Horne A, Jansen F, Kiesel L, et al. ESHRE guideline: endometriosis. Hum Reprod open. 2022;2022(2):hoac009.
Della Corte L, Di Filippo C, Gabrielli O, Reppuccia S, La Rosa VL, Ragusa R, et al. The burden of endometriosis on women’s lifespan: a narrative overview on quality of life and psychosocial wellbeing. Int J Environ Res Public Health. 2020;17(13):4683.
Richardson WS, Carter KM, Fuhrman GM, Bolton JS, Bowen JC. Minimally invasive abdominal surgery. Ochsner J. 2000;2(3):153–7.
Bafort C, Beebeejaun Y, Tomassetti C, Bosteels J, Duffy JM. Laparoscopic surgery for endometriosis. Cochrane Database Syst Reviews. 2020(10).
Patil M Jr, Gharde P, Reddy K, Nayak K. Comparative analysis of laparoscopic versus open procedures in specific general surgical interventions. Cureus. 2024;16(2).
Wright JT. Complications of laparoscopic surgery for endometriosis. Complications in gynecological surgery. Springer; 2008. pp. 34–42.
Quesada J, Härmä K, Reid S, Rao T, Lo G, Yang N, et al. Endometriosis: a multimodal imaging review. Eur J Radiol. 2023;158:110610.
Simko S, Wright KN. The future of diagnostic laparoscopy–cons. Reprod Fertil. 2022;3(2):R91–5.
Parasar P, Ozcan P, Terry KL. Endometriosis: epidemiology, diagnosis and clinical management. Curr Obstet Gynecol Rep. 2017;6:34–41.
Pascoal E, Wessels J, Aas-Eng M, Abrao M, Condous G, Jurkovic D, et al. Strengths and limitations of diagnostic tools for endometriosis and relevance in diagnostic test accuracy research. Ultrasound Obstet Gynecol. 2022;60(3):309–27.
Nisenblat V, Bossuyt PM, Shaikh R, Farquhar C, Jordan V, Scheffers CS, et al. Blood biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Reviews. 1996;2016:5.
Dolińska W, Draper H, Othman L, Thompson C, Girvan S, Cunningham K, et al. Accuracy and utility of blood and urine biomarkers for the noninvasive diagnosis of endometriosis: a systematic literature review and meta-analysis. F&S Reviews; 2022.
Karkia R, Wali S, Payne A, Karteris E, Chatterjee J. Diagnostic accuracy of liquid biomarkers for the noninvasive diagnosis of endometrial cancer: a systematic review and meta-analysis. Cancers. 2022;14(19):4666.
Anastasiu CV, Moga MA, Elena Neculau A, Bălan A, Scârneciu I, Dragomir RM, et al. Biomarkers for the noninvasive diagnosis of endometriosis: state of the art and future perspectives. Int J Mol Sci. 2020;21(5):1750.
Moses AS, Demessie AA, Taratula O, Korzun T, Slayden OD, Taratula O. Nanomedicines for endometriosis: lessons learned from cancer research. Small. 2021;17(7):2004975.
Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MdP, Acosta-Torres LS, et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnol. 2018;16:1–33.
Huseynov E, Khalilov R, Mohamed AJ, NOVEL NANOMATERIALS FOR HEPATOBILIARY, DISEASES TREATMENT AND FUTURE PERSPECTIVES. Adv Biology Earth Sci. 2024;9.
Erdil N, CARDIOVASCULAR, DISEASE, SIGNALING, GENE/CELL THERAPY, AND ADVANCED NANOBIOMATERIALS. Adv Biology Earth Sci. 2024;9.
Rosic G, Selakovic D, Omarova S, CANCER SIGNALING, CELL/GENE THERAPY, DIAGNOSIS AND ROLE OF NANOBIOMATERIALS. Adv Biology Earth Sci. 2024;9.
Yadav VR, Suresh S, Devi K, Yadav S. Novel formulation of solid lipid microparticles of curcumin for anti-angiogenic and anti-inflammatory activity for optimization of therapy of inflammatory bowel disease. J Pharm Pharmacol. 2009;61(3):311–21.
Taheri SL, Rezazadeh M, Hassanzadeh F, Akbari V, Dehghani A, Talebi A, et al. Preparation, physicochemical, and retinal anti-angiogenic evaluation of poloxamer hydrogel containing dexamethasone/avastin-loaded chitosan-N-acetyl-L-cysteine nanoparticles. Int J Biol Macromol. 2022;220:1605–18.
Qiao L, Yang H, Gao S, Li L, Fu X, Wei Q. Research progress on self-assembled nanodrug delivery systems. J Mater Chem B. 2022;10(12):1908–22.
Singh AP, Biswas A, Shukla A, Maiti P. Targeted therapy in chronic diseases using nanomaterial-based drug delivery vehicles. Signal Transduct Target Therapy. 2019;4(1):33.
Marques A, Costa P, Velho S, Amaral M. Functionalizing nanoparticles with cancer-targeting antibodies: a comparison of strategies. J Controlled Release. 2020;320:180–200.
Sahni M, Day ES. Nanotechnologies for the detection and treatment of endometriosis. Front Biomaterials Sci. 2023;2:1279358.
Kalyani T, Sangili A, Nanda A, Prakash S, Kaushik A, Jana SK. Bionanocomposite based highly sensitive and label-free electrochemical immunosensor for endometriosis diagnostics application. Bioelectrochemistry. 2021;139:107740.
Sangili A, Kalyani T, Chen S-M, Nanda A, Jana SK. Label-free electrochemical immunosensor based on one-step electrochemical deposition of AuNP-RGO nanocomposites for detection of endometriosis marker CA 125. ACS Appl Bio Mater. 2020;3(11):7620–30.
Yuxue J, Ran S, Minghui F, Minjia S. Applications of nanomaterials in endometriosis treatment. Front Bioeng Biotechnol. 2023;11:1184155.
Volpini C, Bloise N, Dominoni M, Barra F, Vellone VG, Minzioni P, et al. The nanorevolution in the diagnosis and treatment of endometriosis. Nanoscale. 2023;15(43):17313–25.
Hoyer KJR, Dittrich S, Bartram MP, Rinschen MM. Quantification of molecular heterogeneity in kidney tissue by targeted proteomics. J Proteom. 2019;193:85–92.
Konvalinka A, Scholey JW, Diamandis EP. Searching for new biomarkers of renal diseases through proteomics. Clin Chem. 2012;58(2):353–65.
Zhang T, Duran V, Vanarsa K, Mohan C. Targeted urine proteomics in lupus nephritis–a meta-analysis. Expert Rev Proteomics. 2020;17(10):767–76.
AB S, Srivastava P, Shivaji S. Understanding the pathogenesis of endometriosis through proteomics: recent advances and future prospects. PROTEOMICS–Clinical Appl. 2014;8(1–2):86–98.
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–36.
Li A, Barber RF. Multiple testing with the structure-adaptive Benjamini–Hochberg algorithm. J Royal Stat Soc Ser B: Stat Methodol. 2019;81(1):45–74.
Cho S, Choi YS, Yim SY, Yang HI, Jeon YE, Lee KE, et al. Urinary vitamin D-binding protein is elevated in patients with endometriosis. Hum Reprod. 2012;27(2):515–22.
Faserl K, Golderer G, Kremser L, Lindner H, Sarg B, Wildt L, et al. Polymorphism in vitamin D-binding protein as a genetic risk factor in the pathogenesis of endometriosis. J Clin Endocrinol Metabolism. 2011;96(1):E233–41.
Hwang J-H, Wang T, Lee K-S, Joo J-K, Lee H-G. Vitamin D binding protein plays an important role in the progression of endometriosis. Int J Mol Med. 2013;32(6):1394–400.
Ferrero S, Gillott DJ, Anserini P, Remorgida V, Price KM, Ragni N, et al. Vitamin D binding protein in endometriosis. J Soc Gynecol Investig. 2005;12(4):272–7.
Chen X, Liu H, Sun W, Guo Z, Lang J. Elevated urine histone 4 levels in women with ovarian endometriosis revealed by discovery and parallel reaction monitoring proteomics. J Proteom. 2019;204:103398.
Baek JC, Jo JY, Lee SM, Cho IA, Shin JK, Lee SA, et al. Differences in 25-hydroxy vitamin D and vitamin D-binding protein concentrations according to the severity of endometriosis. Clin Experimental Reproductive Med. 2019;46(3):125.
Costa J, Villa C, Mafra I. 1D-, 2D-Gel electrophoresis, immunoblotting, and enzyme-linked immunosorbent assay (ELISA) for the study of Food allergens. Food Allergens: Methods and Protocols: Springer; 2023. pp. 123–42.
Gh MS, Norouzi F. Guidelines for an optimized differential centrifugation of cells. Biochem Biophys Rep. 2023;36:101585.
Hoofnagle AN, Cobbaert CM, Delatour V, Kelleher NL, Lowenthal MS, Shuford CM. Should LC–MS/MS be the reference measurement procedure to determine protein concentrations in human samples? Clin Chem. 2021;67(3):466–71.
Zhou S, Song Q, Tang Y, Naidong W. Critical review of development, validation, and transfer for high throughput bioanalytical LC–MS/MS methods. Curr Pharm Anal. 2005;1(1):3–14.
Hwang J-H, Lee K-S, Joo J-K, Wang T, Son J-B, Park JH, et al. Identification of biomarkers for endometriosis in plasma from patients with endometriosis using a proteomics approach. Mol Med Rep. 2014;10(2):725–30.
Wölfler MM, Meinhold-Heerlein IM, Söhngen L, Rath W, Knüchel R, Neulen J, et al. Two-dimensional gel electrophoresis in peritoneal fluid samples identifies differential protein regulation in patients suffering from peritoneal or ovarian endometriosis. Fertil Steril. 2011;95(8):2764–8.
Chen S, Liu Y, Zhong Z, Wei C, Liu Y, Zhu X. Peritoneal immune microenvironment of endometriosis: role and therapeutic perspectives. Front Immunol. 2023;14:1134663.
Ferrero S, Gillott DJ, Remorgida V, Anserini P, Price K, Ragni N, et al. Haptoglobin β chain isoforms in the plasma and peritoneal fluid of women with endometriosis. Fertil Steril. 2005;83(5):1536–43.
Ferrero S, Gillott DJ, Remorgida V, Anserini P, Ragni N, Grudzinskas JG. Peritoneal fluid proteome in women with different ASRM stages of endometriosis. Gynecol Endocrinol. 2008;24(8):433–41.
Ferrero S, Gillot DJ, Remorgida V, Grudzinskas JG. Haptoglobin Concentration in Peritoneal Fluid of women with Endometriosis. J Endometr. 2010;2(1):26–32.
Kumpalume JDAPP. 2015Method for the isolation of haptoglobin.
Liau CY, Chang TM, Pan JP, Chen WL, Mao SJ. Purification of human plasma haptoglobin by hemoglobin-affinity column chromatography. J Chromatogr B. 2003;790(1–2):209–16.
Sun L, Huang Y, Zhang Y, Meng Q, Luo J, Fan B, et al. A simple and rapid procedure for purification of haptoglobin from human plasma fraction IV. Artif Cells Blood Substit Biotechnol. 2011;39(2):79–86.
Rauh M. LC–MS/MS for protein and peptide quantification in clinical chemistry. J Chromatogr B. 2012;883:59–67.
Méar L, Com E, Fathallah K, Guillot L, Lavigne R, Guével B, et al. The eutopic endometrium proteome in endometriosis reveals candidate markers and molecular mechanisms of physiopathology. Diagnostics. 2022;12(2):419.
Symons LK, Miller JE, Kay VR, Marks RM, Liblik K, Koti M, et al. The immunopathophysiology of endometriosis. Trends Mol Med. 2018;24(9):748–62.
Goyette J, Geczy CL. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids. 2011;41:821–42.
Laudanski P, Gorodkiewicz E, Ramotowska B, Charkiewicz R, Kuzmicki M, Szamatowicz J. Determination of cathepsins B, D and G concentration in eutopic proliferative endometrium of women with endometriosis by the surface plasmon resonance imaging (SPRI) technique. Eur J Obstet Gynecol Reproductive Biology. 2013;169(1):80–3.
Amaral A, Fernandes C, Morazzo S, Rebordão MR, Szóstek-Mioduchowska A, Lukasik K, et al. The inhibition of cathepsin G on endometrial explants with endometrosis in the mare. Front Veterinary Sci. 2020;7:582211.
Sasamoto N, Ngo L, Vitonis AF, Dillon ST, Missmer SA, Libermann TA, et al. Circulating proteomic profiles associated with endometriosis in adolescents and young adults. Hum Reprod. 2022;37(9):2042–53.
Sikora J, Wróblewska-Czech A, Smycz-Kubańska M, Mielczarek-Palacz A, Cygal A, Witek A, et al. The role of complement components C1q, MBL and C1 inhibitor in pathogenesis of endometriosis. Arch Gynecol Obstet. 2018;297:1495–501.
Agostinis C, Zorzet S, Balduit A, Zito G, Mangogna A, Macor P et al. Complement component 3 expressed by the endometrial ectopic tissue is involved in the endometriotic lesion formation through mast cell activation. bioRxiv. 2020:2020.11. 19.389536.
Agostinis C, Zorzet S, Balduit A, Zito G, Mangogna A, Macor P, et al. The inflammatory feed-forward loop triggered by the complement component C3 as a potential target in endometriosis. Front Immunol. 2021;12:693118.
ISAACSON KB COUTIFARISC, GARCIA C-R, LYTTLE CR. Production and secretion of complement component 3 by endometriotic tissue. J Clin Endocrinol Metabolism. 1989;69(5):1003–9.
Vandooren J, Itoh Y. Alpha-2-macroglobulin in inflammation, immunity and infections. Front Immunol. 2021;12:803244.
Iwasaki S, Kaneda K. Genes relating to biological processes of endometriosis: expression changes common to a mouse model and patients. Drug Res. 2022;72(09):523–33.
Grindheim AK, Saraste J, Vedeler A. Protein phosphorylation and its role in the regulation of annexin A2 function. Biochim et Biophys Acta (BBA)-General Subj. 2017;1861(11):2515–29.
Menzhinskaya IV, Pavlovich SV, Melkumyan AG, Chuprynin VD, Yarotskaya EL, Sukhikh GT. Potential significance of serum autoantibodies to endometrial antigens, α-Enolase and hormones in non-invasive diagnosis and Pathogenesis of endometriosis. Int J Mol Sci. 2023;24(21):15578.
Knudtson JF, McLaughlin JE, Sultana M, Santos MT, Sureshkumar M, Tekmal RR, et al. CD44 variant 6 is involved in the attachment and invasion of endometrial cells to peritoneum. F&S Sci. 2020;1(2):188–94.
Atkins HM, Bharadwaj MS, O’Brien Cox A, Furdui CM, Appt SE, Caudell DL. Endometrium and endometriosis tissue mitochondrial energy metabolism in a nonhuman primate model. Reproductive Biology Endocrinol. 2019;17(1):1–10.
Madanes D, Bilotas MA, Bastón JI, Singla JJ, Meresman GF, Barañao RI, et al. PI3K/AKT pathway is altered in the endometriosis patient’s endometrium and presents differences according to severity stage. Gynecol Endocrinol. 2020;36(5):436–40.
Ding D, Liu X, Duan J, Guo S–W. Platelets are an unindicted culprit in the development of endometriosis: clinical and experimental evidence. Hum Reprod. 2015;30(4):812–32.
Kusama K, Satoyoshi A, Azumi M, Yoshie M, Kojima J, Mizuno Y, et al. Toll-like receptor signalling pathway triggered by inhibition of serpin A1 stimulates production of inflammatory cytokines by endometrial stromal cells. Front Endocrinol. 2022;13:966455.
Liu F, Lv X, Yu H, Xu P, Ma R, Zou K. In search of key genes associated with endometriosis using bioinformatics approach. Eur J Obstet Gynecol Reproductive Biology. 2015;194:119–24.
Shi J-L, Zheng Z-M, Chen M, Shen H-H, Li M-Q, Shao J. IL-17: an important pathogenic factor in endometriosis. Int J Med Sci. 2022;19(4):769.
Yu L, Shen H, Ren X, Wang A, Zhu S, Zheng Y, et al. Multi-omics analysis reveals the interaction between the complement system and the coagulation cascade in the development of endometriosis. Sci Rep. 2021;11(1):11926.
Banerjee S, Xu W, Doctor A, Driss A, Nezhat C, Sidell N et al. TNFα-induced altered miRNA expression links to NF-κB signalling pathway in endometriosis. 2023.
Yin X, Pavone ME, Lu Z, Wei J, Kim JJ. Increased activation of the PI3K/AKT pathway compromises decidualization of stromal cells from endometriosis. J Clin Endocrinol Metabolism. 2012;97(1):E35–43.
Abe W, Nasu K, Tsuno A, Kawano Y, Narahara H. Phosphatidylinositol-3 kinase-akt-mammalian target of rapamycin signalling pathway mediates contractility of human endometriotic stromal cells: a promising new target for the treatment of endometriosis-associated fibrosis. Gynecol Minim Invasive Therapy. 2014;3(4):115–8.
Bora G, Yaba A. The role of mitogen-activated protein kinase signalling pathway in endometriosis. J Obstet Gynecol Res. 2021;47(5):1610–23.
Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodelling in development and disease. Cold Spring Harb Perspect Biol. 2011;3(12):a005058.
Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020;11(1):5120.
Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu X, et al. Extracellular matrix remodelling in tumor progression and immune escape: from mechanisms to treatments. Mol Cancer. 2023;22(1):48.
Lv S-j, Sun J-n, Gan L, Sun J. Identification of molecular subtypes and immune infiltration in endometriosis: a novel bioinformatics analysis and in vitro validation. Front Immunol. 2023;14.
Murakami M, Osuka S, Muraoka A, Hayashi S, Bayasula, Kasahara Y, et al. Effectiveness of NLRP3 inhibitor as a non-hormonal treatment for ovarian endometriosis. Reproductive Biology Endocrinol. 2022;20(1):58.
Zhang M, Shi Z, Peng X, Cai D, Peng R, Lin Y et al. NLRP3 inflammasome-mediated pyroptosis induce Notch signal activation in endometriosis angiogenesis. Mol Cell Endocrinol. 2023:111952.
Irandoost E, Najibi S, Talebbeigi S, Nassiri S. Focus on the role of NLRP3 inflammasome in the pathology of endometriosis: a review on molecular mechanisms and possible medical applications. Naunyn Schmiedebergs Arch Pharmacol. 2023;396(4):621–31.
Agostinis C, Balduit A, Mangogna A, Zito G, Romano F, Ricci G, et al. Immunological basis of the endometriosis: the complement system as a potential therapeutic target. Front Immunol. 2021;11:599117.
Dituri F, Gigante G, Scialpi R, Mancarella S, Fabregat I, Giannelli G. Proteoglycans in cancer: friends or enemies? A special focus on hepatocellular carcinoma. Cancers. 2022;14(8):1902.
Barkovskaya A, Buffone A Jr, Žídek M, Weaver VM. Proteoglycans as mediators of cancer tissue mechanics. Front Cell Dev Biology. 2020;8:569377.
Ji S, Liu Y, Yan L, Zhang Y, Li Y, Zhu Q et al. DIA-based analysis of the menstrual blood proteome identifies association between CXCL5 and IL1RN and endometriosis. J Proteom. 2023:104995.
Višnić A, Jurešić GČ, Domitrović R, Klarić M, Šepić TS, Barišić D. Proteins in urine–possible biomarkers of endometriosis. J Reprod Immunol. 2023;157:103941.
Penariol LB, Thomé CH, Tozetti PA, Paier CR, Buono FO, Peronni KC, et al. What do the transcriptome and proteome of menstrual blood-derived mesenchymal stem cells tell us about endometriosis? Int J Mol Sci. 2022;23(19):11515.
Giuseppe G, Domenico M, Domenico R, Francesca M, Alfredo P, Riccardo M, et al. Identification of novel putative urinary markers of endometriosis by high-resolution quantitative proteomics. Biomedical J Sci Tech Res. 2020;28(2):21475–81.
Manousopoulou A, Hamdan M, Fotopoulos M, Garay-Baquero DJ, Teng J, Garbis SD, et al. Integrated Eutopic Endometrium and non‐depleted serum quantitative proteomic analysis identifies candidate serological markers of endometriosis. PROTEOMICS–Clinical Appl. 2019;13(3):1800153.
Grande G, Vincenzoni F, Milardi D, Pompa G, Ricciardi D, Fruscella E, et al. Cervical mucus proteome in endometriosis. Clin Proteomics. 2017;14:1–11.
Zhao Y, Liu Y-N, Li Y, Tian L, Ye X, Cui H, et al. Identification of biomarkers for endometriosis using clinical proteomics. Chin Med J. 2015;128(04):520–7.
Dutta M, Subramani E, Taunk K, Gajbhiye A, Seal S, Pendharkar N, et al. Investigation of serum proteome alterations in human endometriosis. J Proteom. 2015;114:182–96.
Wang L, Liu H, Shi H, Lang J, Sun W. Urine peptide patterns for noninvasive diagnosis of endometriosis: a preliminary prospective study. Eur J Obstet Gynecol Reproductive Biology. 2014;177:23–8.
Williams KE, Miroshnychenko O, Johansen EB, Niles RK, Sundaram R, Kannan K, et al. Urine, peritoneal fluid and omental fat proteomes of reproductive age women: endometriosis-related changes and associations with endocrine disrupting chemicals. J Proteom. 2015;113:194–205.
Long X, Jiang P, Zhou L, Zhang W. Evaluation of novel serum biomarkers and the proteomic differences of endometriosis and adenomyosis using MALDI-TOF–MS. Arch Gynecol Obstet. 2013;288:201–5.
Fassbender A, Waelkens E, Verbeeck N, Kyama CM, Bokor A, Vodolazkaia A, et al. Proteomics analysis of plasma for early diagnosis of endometriosis. Obstet Gynecol. 2012;119(2 Part 1):276–85.
El-Kasti MM, Wright C, Fye HK, Roseman F, Kessler BM, Becker CM. Urinary peptide profiling identifies a panel of putative biomarkers for diagnosing and staging endometriosis. Fertil Steril. 2011;95(4):1261–6. e6.
Seeber B, Sammel MD, Fan X, Gerton GL, Shaunik A, Chittams J, et al. Proteomic analysis of serum yields six candidate proteins that are differentially regulated in a subset of women with endometriosis. Fertil Steril. 2010;93(7):2137–44.
Tokushige N, Markham R, Crossett B, Ahn SB, Nelaturi VL, Khan A, et al. Discovery of a novel biomarker in the urine in women with endometriosis. Fertil Steril. 2011;95(1):46–9.
Jing J, Qiao Y, Suginami H, Taniguchi F, Shi H, Wang X. Two novel serum biomarkers for endometriosis screened by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry and their change after laparoscopic removal of endometriosis. Fertil Steril. 2009;92(4):1221–7.
Zhang H, Feng J, Chang X-h, Li Z-x, Wu X-y, Cui H. Effect of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry on identifing biomarkers of endometriosis. Chin Med J. 2009;122(04):373–6.
Liu H, Zheng Y, Zhang J, Leng J, Sun D, Liu Z, et al. Establishment of endometriosis diagnostic model using plasma protein profiling. Zhonghua Fu Chan Ke Za Zhi. 2009;44(8):601–4.
Wölfler MM, Schwamborn K, Otten D, Hornung D, Liu H, Rath W. Mass spectrometry and serum pattern profiling for analysing the individual risk for endometriosis: promising insights? Fertil Steril. 2009;91(6):2331–7.
Wang L, Zheng W, Mu L, Zhang S-Z. Identifying biomarkers of endometriosis using serum protein fingerprinting and artificial neural networks. Int J Gynecol Obstet. 2008;101(3):253–8.
Liu H, Lang J, Zhou Q, Shan D, Li Q. Detection of endometriosis with the use of plasma protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Fertil Steril. 2007;87(4):988–90.
Acknowledgements
Not applicable.
Funding
NA.
Author information
Authors and Affiliations
Contributions
G.G.A. initially began the review and wrote the protocol with help from W.C.C. and Z.T. G.G.A. and B.A.K. performed the data extraction and quality assessment for the selected articles. The analysis was carried out by G.G.A. and W.L. G.G.A. wrote the first draft of the manuscript with the help of W.C.C., Z.T., C.E.C.W., L.W.F., F.L.W.Y. and W.L., who provided feedback on the review and modifications. All authors contributed to and approved the final version of this article.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
: Figure S1. QUADAS-2 tool: The distribution of risk-of-bias (A) and applicability (B) judgments within each bias domain. Figure S2. Network of enriched GO terms in peripheral blood (plasma): (a) biological process, (b) cellular component and (c) molecular function. Figure S3. Network of enriched GO terms in peripheral blood (serum): (a) biological process, (b) cellular component and (c) molecular function. Figure S4. Network of enriched GO terms in menstrual blood. (a) biological process (b) cellular component and (c) molecular function. Figure S5. Network of enriched GO terms in urine: (a) biological process, (b) cellular component and (c) molecular function. Figure S6. GO term analysis of DEPs in plasma, serum, menstrual blood, and urine from patients with endometriosis
Supplementary Material 2
: Table S1. List of differentially expressed proteins
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Azeze, G.G., Wu, L., Alemu, B.K. et al. Proteomics approach to discovering non-invasive diagnostic biomarkers and understanding the pathogenesis of endometriosis: a systematic review and meta-analysis. J Transl Med 22, 685 (2024). https://doi.org/10.1186/s12967-024-05474-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-024-05474-3